————————————-
Vaccine Types
Scientific research has led to the development of numerous types of vaccines that safely elicit immune responses that protect against infection, and researchers continue to investigate novel vaccine strategies for prevention of existing and emerging infectious diseases. Recent decades have brought major advances in understanding the complex interactions between the microbes that cause disease and their human hosts. These insights, as well as advances in laboratory techniques and technologies, have aided the development of new types of vaccines.
Whole-Pathogen Vaccines
Subunit Vaccines
Nucleic Acid Vaccines
https://www.niaid.nih.gov/research/vaccine-types
————————————-
Understanding the nanotechnology in COVID-19 vaccines
2021
https://fightingmonarch.files.wordpress.com/2021/07/the-enemy-man-splains-the-nanotech-in-the-vaccines.pdf
————————————-
Role of nanotechnology behind the success of mRNA vaccines for
COVID-19
2021
https://fightingmonarch.files.wordpress.com/2021/08/role-of-nanotechnology-behind-the-success-of-mrna-vaccines-for-covid-19.pdf
————————————-
How Nanotechnology Helped Create mRNA COVID-19 Vaccines
Dec 4 2020
https://www.azonano.com/news.aspx?newsID=37659
————————————-
Nano-Niclosamide Has potential to Treat COVID-19
May 6, 2021
https://www.medindia.net/news/nano-niclosamide-has-potential-to-treat-covid-19-201075-1.htm
————————————-
Slight pH adjustment may turn a metabolic inhibiting drug into promising COVID-19 treatment
Jan 7 2022
https://www.news-medical.net/news/20220107/Slight-pH-adjustment-may-turn-a-metabolic-inhibiting-drug-into-promising-COVID-19-treatment.aspx
Mechanical engineering and materials science professor David Needham has shown that a slight increase in solution pH might be all it takes to turn a metabolic inhibiting drug, traditionally used to treat gut parasites, into a promising prophylactic/preventative nasal spray and early treatment throat spray for COVID-19.
The results appear online on December 28 in the journal Pharmaceutical Research.
Since 1958, niclosamide has been used to treat gut parasite infections in humans, pets and farm animals. Delivered as oral tablets, the drug kills the parasites on contact by inhibiting their crucial metabolic pathway and shutting down their energy supply.
In recent years, however, researchers have been testing niclosamide’s potential to treat a much wider range of diseases, such as many types of cancer, metabolic diseases, rheumatoid arthritis and systemic sclerosis. Recent laboratory studies in cells have also shown the drug to be a potent antiviral medication, inhibiting a virus’s ability to cause disease by targeting the energy supply of the host cell that the virus co-opts for its self-replication.
Niclosamide primarily acts upon host cell’s mitochondria, which are like energy-producing batteries of the cell. The drug prevents the cell from producing its main energy molecule, adenosine 5′-triphosphate, or ATP. Without the infected cell’s energy supply, the virus has trouble replicating viable copies of itself to cause further infections. These effects are reversible and do not result in any cell death.
“Niclosamide turns down the dimmer switch on a cell’s energy and essentially puts the virus in lockdown,” said Needham, the sole author of the new study. When used in conjunction with vaccines, masking and other recommended mitigation measures for COVID prevention, the new niclosamide solution holds potential as an adjunct strategy, he said. “This development could enable safe and effective nose and throat sprays that provide additional protection behind the mask.”
————————————-
Hydrotalcite–Niclosamide Nanohybrid as Oral Formulation towards SARS-CoV-2 Viral Infections
2021 May 19
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8160721/
————————————-
Scientists Find New Possible Weapon Against COVID-19 In Synthetic Nano-Bodies
Nov 15, 2020
Highlights
A team of scientists from EMBL Hamburg have now identified synthetic mini-antibodies which can potentially be used to battle against COVID-19
The group, led by Dr Christian Low, has found a new method to block the SARS-CoV-2 from infecting human cells
Now published in Nature Communications, The method makes use of synthetic nanobodies, called sybodies, that prevent the virus from binding onto the human cells and hence from getting a person infected with the virus
Sybodies are the synthetic replicas of nanobodies, small antibodies found in camels and llamas, explains an ANI report. These antibodies are effective against viruses due to their high stability and small size.
https://www.indiatimes.com/technology/news/covid-19-synthetic-nano-bodies-526936.html
————————————-
Nano-curcumin therapy, a promising method in modulating inflammatory cytokines in COVID-19 patients
2020 Oct 20
https://pubmed.ncbi.nlm.nih.gov/33129099/
————————————-
Nanotech May Help Fight ‘Cytokine Storm’ of COVID
April 28, 2020
https://www.webmd.com/lung/news/20200428/nanotech-may-help-fight-cytokine-storm-of-covid-19#1
————————————-
Decoy nanoparticles protect against COVID-19 by concurrently adsorbing viruses and inflammatory cytokines
2020 Oct 6
Abstract
The COVID-19 pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has highlighted the urgent need to rapidly develop therapeutic strategies for such emerging viruses without effective vaccines or drugs. Here, we report a decoy nanoparticle against COVID-19 through a powerful two-step neutralization approach: virus neutralization in the first step followed by cytokine neutralization in the second step. The nanodecoy, made by fusing cellular membrane nanovesicles derived from human monocytes and genetically engineered cells stably expressing angiotensin converting enzyme II (ACE2) receptors, possesses an antigenic exterior the same as source cells. By competing with host cells for virus binding, these nanodecoys effectively protect host cells from the infection of pseudoviruses and authentic SARS-CoV-2. Moreover, relying on abundant cytokine receptors on the surface, the nanodecoys efficiently bind and neutralize inflammatory cytokines including interleukin 6 (IL-6) and granulocyte-macrophage colony-stimulating factor (GM-CSF), and significantly suppress immune disorder and lung injury in an acute pneumonia mouse model. Our work presents a simple, safe, and robust antiviral nanotechnology for ongoing COVID-19 and future potential epidemics.
Polymer-based nano-therapies to combat COVID-19 related respiratory injury: progress, prospects, and challenges
2021 Apr 14
https://pubmed.ncbi.nlm.nih.gov/33787467/
————————————-
Developments in Nano-materials and Analysing its role in Fighting COVID-19
2021
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8106907/
————————————-
Antidote for Spike Proteins & COVID19 Vaccination? Fennel, Star Anise, Shikimic acid, Pine Tree Needle Turpentine & NANO SOMA
May 16, 2021
https://www.survivethenews.com/antidote-spike-proteins-covid19-vaccination-fennel-star-anise-pine-needle-tea-turpentine-nano-soma/
————————————-
OPERATION COVID-19 & the Nanotech Agenda
November 29, 2020
https://stateofthenation.co/?p=39099
————————————-
Nanotechnology shown to slow spread of COVID-19 virus in lung and white blood cells, study shows
Jul 03, 2020
SAN DIEGO, California — A promising technology slowed the spread of SARS-CoV-2, the virus that causes COVID-19, in cell cultures, researchers at the University of California San Diego and Boston University found in lab experiments.
The United States led the world in coronavirus cases with 2.7 million confirmed Thursday, according to data maintained by Johns Hopkins University.
Engineers at UC-San Diego coated tiny nanoparticles made of polymer with lung and white blood cell membranes, disguising them as human cells to the virus.
The membranes covering the nanoparticles had the same external receptors and proteins that the virus uses to enter the human lung and white blood cells. The nanoparticles fooled the SARS-CoV-2 virus into thinking they were human cells and the virus bound onto them. Once attached to the nanoparticles, the virus could no longer enter a human cell or reproduce.
These lung cells and white blood cell nanoparticles blocked almost 90 percent of the virus’ ability to enter human cells, reproduce and create more virus in lab dish experiments, researchers out of UC San Diego and Boston University published last month in Nano Letters.
Nanoparticles were first masked as human cells, like red blood cells, more than a decade ago at UC San Diego’s Jacobs School of Engineering. They can also be used to extract oil or toxins from water or an oil spill. They have to be masked to be used in the body because the immune system attacks foreign objects. They have been dubbed nanosponges by researchers because of their ability to soak up pathogens or toxins.
Researchers at UC-San Diego will work next to see how well the COVID-19 nanosponges work in animals, and potentially, in humans.
https://www.cleveland.com/metro/2020/07/nanotechnology-shown-to-slow-spread-of-covid-19-virus-in-lung-and-white-blood-cells-study-shows.html
————————————-
Nanotechnology in Washable and Reusable Face Masks
Jul 16 2020
https://www.azonano.com/article.aspx?ArticleID=5529
Reusable Nanotechnology-Based Face masks
Researchers at the Korea Advanced Institute of Science and Technology (KAIST) has recently announced its development of a reusable nano-filtered facemask.
Professor Il-Doo Kim of KAIST claims that this mask retains its filtering efficiency and sturdy frame even after 20 washes.
The nano filters used in the development of the facemask can filter out the finest dust particles. This reusable nano-filtered facemask would be an economical option for daily usage as disposable masks often end up being very expensive when purchasing it on a regular basis.
This type of facemask could also relieve the challenges arising from the scarcity of face masks. The research team at KAIST is awaiting approval from the Ministry of Food and Drug Safety for launching their product in the commercial market.
The structure and alignment of nanoparticles in the nano filters of this facemask make it better than conventional disposable face masks. These commonly available face masks have to be disposed of after one use as they lose their electrostatic function when exposed to water molecules. Therefore, they cannot maintain proper air filtration.
Following the insulation block electrospinning process, Professor Kim at KAIST developed the facemask with the help of orthogonal nanofibers. The structure of the nanofibers minimizes air filter pressure and enhances the filtration process.
The nanofiber filter of the face masks is water-resistant and has a 94% filtration efficiency in 20 repeated bactericidal tests.
It could also retain the structure of the nano-membrane, even after it was hand washed repeatedly.
No deformations in the nanomembrane were observed, even after it was soaked in ethanol for more than three hours.
The research team said that this facemask could be reusable for around one month and the inner filter could be replaced if required.
Professor Kim recently founded a startup company called “Kim Il-Doo Research Institute”, which can produce around 1,500 nanofiber filters per day.
Development of Face Masks using Breathable Nanocellulose
Dr. Thomas Rainey and his research team at Queensland University of Technology (QUT) have developed nanotechnology-based biodegradable face masks.
The facemask filter is made up of a cellulose nanofiber component. These nanofibers are produced from waste plant material such as agricultural waste, sugarcane, bagasse, etc., and can be easily biodegraded.
The breathable nanocellulose material filters out 100-nanometer particles, which is the size of several viruses.
Reusable Face Masks with Antimicrobial Property
An Israeli startup Sonovia designed reusable face masks with antimicrobial properties. This is made up of cotton incorporated with metal-oxide nanoparticles. The usage of zinc-oxide nanoparticles showed the efficiency of killing germs and the capacity to last for more than 100 washes.
At present, these masks are highly efficient in blocking almost 100% of bacteria and 98% of 5-micron particles. However, the company aims high, i.e., 3 microns filtration. As their long-term plan, the research team envisioned a future model that will have the option to incorporate a 0.2-micron filter to filter high-velocity aerosols.
These masks are commonly referred to as SonoMask, and have been commercially available since March 2020.
Initially, the company had donated large numbers of masks to hospitals and nonprofit charitable organizations in many countries, including Israel and Germany. Sonovia produces around 20,000 masks each day in the factory located in the northern part of Israel and sells it to retailers and medical institutions all around the world.
Sonovia research scientist Jason Migdal said that Sonovia would soon be listed on the New York Stock Exchange, and it is aiming to expand its technological application from just face masks to all PPE by the end of 2020.
————————————-
With nano-diamonds and salt, researchers race to design a face mask that kills the coronavirus
March 29, 2020
https://fortune.com/2020/03/29/coronavirus-face-mask-shortage-new-design-covid-19/
————————————-
Salt-coated masks and air filters to potentially slow the spread of COVID-19
2020
https://hospitalnews.com/salt-coated-masks-and-air-filters-to-potentially-slow-the-spread-of-covid-19/
————————————-
Nano-Silver Masks May Be Helping to Control Covid in Vietnam
Jul 28, 2020
https://www.registerguard.com/ZZ/news/20200728/nano-silver-masks-may-be-helping-to-control-covid-in-vietnam
————————————-
Know the Health Risks Before Investing in an Antimicrobial Nano-Silver Mask
November 23, 2020
https://www.centerforfoodsafety.org/blog/6201/know-the-health-risks-before-investing-in-an-antimicrobial-nano-silver-mask-and-what-to-buy-instead
————————————-
Personalized Reusable Face Masks with Smart Nano-Assisted Destruction of Pathogens for COVID-19: A Visionary Road
07 December 2020
https://chemistry-europe.onlinelibrary.wiley.com/doi/10.1002/chem.202004875
————————————-
Nanotechnology and Nanomaterials Solutions for COVID-19: Diagnostic Testing, Antiviral and Antimicrobial Coatings and Surfaces, Air-Borne Filtration, Facemasks, PPE, Drug Delivery and Therapeutics
May 2020
https://www.researchandmarkets.com/reports/5023699/nanotechnology-and-nanomaterials-solutions-for
————————————-
Israeli startup says its nanotech masks and robes may block coronavirus
6 February 2020
Sonovia’s textiles are resistant to bacteria and fungus; firm hopes its tech will also work against viruses; has sent samples for testing to China
An Israeli startup, Sonovia Ltd., says it may have the ability to create virus resistance masks and textiles to help combat the coronavirus by using a nanotechnology process it developed to impregnate textiles with antifungal and antibacterial chemicals.
The firm says that using a patented nanotechnology process developed at Israel’s Bar-Ilan University it has managed to create masks and protective textile equipment that have proven effective in blocking the penetration of bacteria and fungus. Now, Jason Migdal, in charge of business development at the firm, says he believes the same method could also be beneficial to halt viruses, and could be effective in helping halt the spread of the deadly coronavirus.
Migdal said the startup has sent textile samples impregnated with zinc using the same method to the Chinese Academy of Sciences in Beijing, for its lab to test the efficacy of the method against viruses.
Studies have shown that nanoparticles of zinc, silver and graphite are all viral inhibitors, Migdal told The Times of Israel in a phone interview, which is why the firm “has good reason to believe” that its process could also be useful in fighting viruses.
The Chinese lab will follow a European protocol to test the fabric’s anti-viral activity, Migdal said.
The virus that started in China has infected at least 28,000 people globally, and more than 560 have died. The World Health Organization has declared the outbreak a global emergency and has warned governments to prepare for “domestic outbreak control” if the disease were to spread in their countries.
Sonovia has developed an ultrasonic fabric finishing technology for the mechanical impregnation of zinc oxite nanoparticles — which are known to be strong anti-microbial and anti-viral agents — into textiles in a permanent manner.
The process uses soundwaves in water that break down the zinc into nanoparticles that are formed within bubbles of air. When the bubbles explode, they create tiny jet streams of liquid that force the nanoparticles of zinc into the surface of the fabric, “for long-term durability,” Migdal said.
The impregnated textile is able to maintain its anti-pathogen activity at up to 100 washes at 75° Celsius and 65 washes at 92° Celsius, he said.
“We have demonstrated clear bactericidal activity against a broad spectrum of infections in laboratory tests,” he said. A pilot study at a European hospital showed that when the impregnated fabrics were used in protective clothing, there was a “significant reduction” of infections.
“There was less bacteria in the ears and in the mouth,” he said.
https://www.timesofisrael.com/israeli-startup-says-its-nanotech-masks-robes-may-block-coronavirus/
————————————-
Lab tests suggest Israeli-made face mask eliminates over 99% of coronavirus
DECEMBER 27, 2020
Sonovia’s reusable anti-viral masks are coated in zinc oxide nano-particles that destroy bacteria, fungi and viruses, which it says can help stop the spread of the coronavirus.
https://www.jpost.com/health-science/israeli-made-mask-eliminates-over-99-percent-of-coronavirus-lab-tests-suggest-644434
————————————-
Fact Check- Fibres in COVID-19 test swabs and face masks are not alive
May 4, 2021
https://www.reuters.com/article/factcheck-fibres-alive-idUSL1N2MR1NW
————————————-
Are There Black Worms In Your Imported Face Masks? Here’s The Truth
April 2021
https://sg.style.yahoo.com/black-worms-imported-face-masks-091811290.html
————————————-
New coating technology uses ‘nanoworms’ to kill COVID-19
September 15, 2021

An antiviral surface coating technology sprayed on face masks could provide an extra layer of protection against COVID-19 and the flu.
The coating developed at The University of Queensland has already proven effective in killing the virus that causes COVID-19, and shows promise as a barrier against transmission on surfaces and face masks.
UQ’s Australian Institute for Bioengineering and Nanotechnology researcher Professor Michael Monteiro said the water-based coating deployed worm-like structures that attack the virus.
“When surgical masks were sprayed with these ‘nanoworms,” it resulted in complete inactivation of the Alpha variant of SARS-CoV-2 and influenza A,” Professor Monteiro said.
The coating was developed with Boeing as a joint research project, and was tested at the Peter Doherty Institute for Infection and Immunity at The University of Melbourne.
“These polymer ‘nanoworms’ rupture the membrane of virus droplets transmitted through coughing, sneezing or saliva and damage their RNA,” Professor Monteiro said.
————————————-
COVID-19 Nasal Swab Test Led To Cerebrospinal Fluid Leak
October 5, 2020
https://www.forbes.com/sites/ninashapiro/2020/10/05/covid-19-nasal-swab-test-led-to-spinal-fluid-leak/?sh=36bae9db35e9
————————————-
Cribriform Plate Injury After Nasal Swab Testing for COVID-19
September 9, 2021
https://jamanetwork.com/journals/jamaotolaryngology/fullarticle/2784128
————————————-
Complications of nasal SARS-CoV-2 testing: a review
2021
https://jim.bmj.com/content/early/2021/08/03/jim-2021-001962?utm_term=jim&utm_content=BMJUK_TMD_CM_2021&utm_campaign=usage&utm_medium=trendmd&utm_source=trendmd
————————————-
Covid-19 swab tests made by Randox and given to thousands of Britons have been HALTED over fears they are unsafe
16 July 2020
https://www.dailymail.co.uk/news/article-8530333/Coronavirus-swab-tests-unsafe-halted.html
Coronavirus tests given out to thousands of Britons have been retracted over fears they are not safe, the Government has announced.
The Department of Health has instructed care homes and members of the public to immediately stop using the kits produced by Randox Laboratories.
Health bosses have refused to reveal the number of swab kits affected, or how many Britons used them before they were pulled.
Care homes and public told to immediately stop using kits made by Randox labs
Health bosses have refused to reveal the number of swab kits that are affected
But Northern Irish firm is second largest provider of UK’s coronavirus tests
But the Northern Irish firm won a £133million contract to carry out at-home Covid-19 tests and ones administered at drive-through centres and care homes.
Under the deal, the swab kits are posted back to Randox for it to process and see if someone has the virus.
So tests made by the manufacturer likely account for a huge chunk of the 150,000 swabs being carried out every day in Britain.
MailOnline understands safety concerns were raised yesterday when it emerged that one of Randox’s suppliers had not provided safety assurance documents.
Health Secretary Matt Hancock told the House of Commons tonight that physical checks were carried out on the swabs which ‘were not up to the standards that we expect…’
Randox was awarded the contract by the Department of Health to help make testing kits that the government could use to ramp up its capacity to carry out 100,000 swabs each day back in April.
The Government was criticised for the Randox deal after it came to light that Tory MP Owen Paterson receives a six-figure salary from the firm to act as a consultant.
Randox’s tests can produce results in a matter of hours and machines that analyse the swabs can process 54 samples at once.
The Telegraph reports that this is not the first time there have been problems with Randox tests during the UK’s fight against Covid-19.
In May, a machine at the firm’s lab in County Antrim stopped working and the UK was forced to send tens of thousands of samples to a lab in the US.
But nearly 30,000 of the swabs had to be discarded because they took to long to arrive.
Samples have to be tested within 72 hours of the test being taken, which means that any delay in their processing could leave people with symptoms unsure if they have the virus.
In a statement, Randox said: ‘As an immediate precautionary measure we have temporarily suspended distribution of home sample collection kits using one particular batch/supplier of swabs.
‘This is a temporary measure and does not apply to our private business which uses a different supplier of swabs.’
————————————-
Covid Home Test Kit Contains Deadly Chemical Sodium Azide
December 24, 2021
https://infoarmed.com/covid-home-test-kit-contains-deadly-chemical-sodium-azide/
————————————-
Lethal Drug Included In Over The Counter Covid Test Kits
Dec 20, 2021
https://banned.video/watch?id=61c0b341724c932b860a7175
————————————-
Antigen Test, Covid-19, Anterior Nasal Swabs (COVAG025-U) (230$)
https://www.mdsupplies.com/medical-supplies-GenBody-Antigen+Test+Covid-19+Anterior+Nasal+Swabs-XGJCGLPAU1.html
– Requires a CLIA Certificate Number
Rapid Diagnostic Test for the Detection of SARS-CoV-2 Antigen – Results in 15 minutes
GenBody COVID-19 Antigen Rapid Test Kit
Anterior Nasal Swab
Expiry: One year from the date of manufacturing
Detection kit for SARS-CoV-2 antigen in nasopharyngeal or anterior nasal swab specimens.
Rapid detection of SARS-CoV-2 will play a key role in the global spread of the virus.
Affordable and sensitive test that does not require an additional reader, with a processing time of 15-20 minutes.
GenBody COVID-19 Antigen Rapid Test Kit
Anterior Nasal Swab
Features
– Detects SARS-CoV-2 nucleocapsid protein antigen
– Rapid results in 15-20 minutes
– Anterior nasal swab specimen collection
– Identifies acute infection with a 92.31% sensitivity and 99.04% specificity
– For use in patient care settings operating under a
– CLIA Certificate of Waiver, Certificate of Compliance or Certificate of Accreditation.
– The confirmed LoD for the GenBody COVID-19 Ag is 1.11 x 10 TCID50/mL.
MATERIALS PROVIDED
Kit Component: Kit Component, Device
Quantity: Twenty-five (25) single use Test Devices
Description: Individually pouched devices with a desiccant. Test Device contains one reactive test strip. The test strip contains a membrane coated with mouse anti-SARS-CoV-2 NP antibodies for the test line and mouse anti-Nus tag antibodies for the control line, and a conjugate pad impregnated with Mouse anti-SARS-CoV-2 NP antibodies and recombinant Nus tag antigens
Kit Component: Extraction Solution
Quantity: Two (2) bottles containing 9 mL of Extraction Solution
Description: Buffer with detergent and preservative (< 0.1% sodium azide)
Kit Component: Extraction Tube
Quantity: Twenty-five (25) single use tubes
Description: Flexible plastic tube for extraction of sample
Kit Component: Dropper Tips
Quantity: Twenty-five (25) single use dropper tips
Description: Disposable of the Extraction Tube for dispensing the extracted sample
Kit Component: Sterilized Nasopharyngeal or Anterior Nasal Swabs
Quantity: Twenty-five (25) single use specimen sampling swabs
Description: Swab for nasopharyngeal or anterior nasal sample collection with a flexible/breakable handle
Kit Component: External Positive Control Swab
Quantity: One (1) single use swab
Description: Individually pouched swab coated with noninfectious recombinant SARS-CoV-2 protein antigen on the head
Kit Component: External Negative Control Swab
Quantity: One (1) single use swab
Description: Individually pouched swab coated with buffer on the head
————————————-
Leading COVID Test Firm is Planning to Sell Swabs Containing Customer’s DNA
November 15, 2021
https://timcast.com/news/leading-covid-test-firm-is-planning-to-sell-swabs-containing-customers-dna/
————————————-
China’s Nightmarish New Bio Weapon Targets Race and Ethnicity
Oct 8, 2021
https://www.youtube.com/watch?v=biNxl7tiVSY
————————————-
Coronavirus test swabs aren’t your standard Q-tips, and they’re running out as testing ramps up
2020
The two top makers of the highly specialized swabs used to test patients for the novel coronavirus are straining to keep up with the demand, even as both the Italian and U.S. governments are working with them to increase production.
The nasopharyngeal swabs required for the coronavirus tests are quite different from your standard Q-tips – and the exploding need for them has created a bottleneck in the soaring demand for diagnoses.
The swabs have to be long and skinny enough to get to the nasopharynx, the upper part of the throat, behind the nose. They must be made of synthetic fiber and cannot have a wooden shaft. Nor can they contain calcium alginate, a substance typically used for swab tips in wound care, as that can kill the virus, according to the Centers for Disease Control and Prevention.
https://www.usatoday.com/story/news/health/2020/03/18/coronavirus-testing-nasopharyngeal-swabs-running-out-tests-ramp-up/2863270001/
————————————-
ABRASIVE “PORCUPINE” SWABS: DR. ANTONIETTA GATTI’S RESEARCH ON COVID SWAB ELEMENTAL COMPOSITION (Controversial)
August 24th, 2021
https://www.bitchute.com/video/2piOlZuvOFr5/
————————————-
Do the PCR test swabs contain “star-shaped micro-devices” that are secretly vaccinating Covid bioweapon victims?!
February 11, 2021
https://stateofthenation.co/?p=52009
————————————-
No, nasal swabs for a corona test are not like a “punishment for slaves” in ancient Egypt
2020
With the image of a historical situation in Egypt, it is suggested on Facebook that a nasopharynx swab for a corona test is similar to a punishment method for slaves. This is wrong, the picture shows a medical eye treatment.

————————————-
Transnasal excerebration surgery in ancient Egypt
2012 Jan 6
Abstract
Ancient Egyptians were pioneers in many fields, including medicine and surgery. Our modern knowledge of anatomy, pathology, and surgical techniques stems from discoveries and observations made by Egyptian physicians and embalmers. In the realm of neurosurgery, ancient Egyptians were the first to elucidate cerebral and cranial anatomy, the first to describe evidence for the role of the spinal cord in the transmission of information from the brain to the extremities, and the first to invent surgical techniques such as trepanning and stitching. In addition, the transnasal approach to skull base and intracranial structures was first devised by Egyptian embalmers to excerebrate the cranial vault during mummification. In this historical vignette, the authors examine paleoradiological and other evidence from ancient Egyptian skulls and mummies of all periods, from the Old Kingdom to Greco-Roman Egypt, to shed light on the development of transnasal surgery in this ancient civilization. The authors confirm earlier observations concerning the laterality of this technique, suggesting that ancient Egyptian excerebration techniques penetrated the skull base mostly on the left side. They also suggest that the original technique used to access the skull base in ancient Egypt was a transethmoidal one, which later evolved to follow a transsphenoidal route similar to the one used today to gain access to pituitary lesions.
————————————-
Screaming in shock as they are cruelly restrained as tubes are shoved up their noses: The monkeys taken to their deaths by Air France – the only major airline still transporting animals for experiments
16 March 2015
The monkeys are seen having tubes forced up their noses and down their throats to sedate them before the testing
Shocked: The footage shows the monkeys being tested on


————————————-
Activists Unveil Graphic Video of Monkey Brain Implants at Cybernetics Institute
September 12, 2014
https://www.vice.com/en/article/gvyd77/soko-animal-rights-shows-the-cruel-fate-of-tbingen-laboratory-monkeys
————————————-
Rapid Detection of COVID-19 Causative Virus (SARS-CoV-2) in Human Nasopharyngeal Swab Specimens Using Field-Effect Transistor-Based Biosensor
2020 Apr 20
Abstract
Coronavirus disease 2019 (COVID-19) is a newly emerging human infectious disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2, previously called 2019-nCoV). Based on the rapid increase in the rate of human infection, the World Health Organization (WHO) has classified the COVID-19 outbreak as a pandemic. Because no specific drugs or vaccines for COVID-19 are yet available, early diagnosis and management are crucial for containing the outbreak. Here, we report a field-effect transistor (FET)-based biosensing device for detecting SARS-CoV-2 in clinical samples. The sensor was produced by coating graphene sheets of the FET with a specific antibody against SARS-CoV-2 spike protein. The performance of the sensor was determined using antigen protein, cultured virus, and nasopharyngeal swab specimens from COVID-19 patients. Our FET device could detect the SARS-CoV-2 spike protein at concentrations of 1 fg/mL in phosphate-buffered saline and 100 fg/mL clinical transport medium. In addition, the FET sensor successfully detected SARS-CoV-2 in culture medium (limit of detection [LOD]: 1.6 × 101 pfu/mL) and clinical samples (LOD: 2.42 × 102 copies/mL). Thus, we have successfully fabricated a promising FET biosensor for SARS-CoV-2; our device is a highly sensitive immunological diagnostic method for COVID-19 that requires no sample pretreatment or labeling.

Schematic diagram of COVID-19 FET sensor operation procedure. Graphene as a sensing material is selected, and SARS-CoV-2 spike antibody is conjugated onto the graphene sheet via 1-pyrenebutyric acid N-hydroxysuccinimide ester, which is an interfacing molecule as a probe linker.
————————————-
Diagnostic biosensor quickly detects SARS-CoV-2 from nasopharyngeal swabs
April 20, 2020
https://www.sciencedaily.com/releases/2020/04/200420145029.htm
————————————-
Quantitative proteomic dataset from oro- and naso-pharyngeal swabs used for COVID-19 diagnosis: Detection of viral proteins and host’s biological processes altered by the infection
2020 Aug 5
https://pubmed.ncbi.nlm.nih.gov/32835036/
————————————-
Nasopharyngeal Swabs Are More Sensitive Than Oropharyngeal Swabs for COVID-19 Diagnosis and Monitoring the SARS-CoV-2 Load
2020
https://pubmed.ncbi.nlm.nih.gov/32626720/
————————————-
Early antiviral response in the nose may determine the course of COVID-19
July 29, 2021
https://imes.mit.edu/early-antiviral-response-in-the-nose-may-determine-the-course-of-covid-19/
————————————-
Please remain calm while the robot swabs your nose
Aug 24, 2020
A medtech startup wants to automate COVID-19 swabs
https://www.theverge.com/2020/8/24/21377011/robot-nasal-swab-machine-autonomous-covid-19-test-brain-navi
————————————-
BRAIN BUSTER: Coronavirus swab test poked so far up woman’s nose it caused her brain to LEAK
1 Oct 2020
A CORONAVIRUS swab test was poked so far up a woman’s nose it caused her brain to LEAK, doctors have revealed.
The patient went to see a doctor after experiencing a metallic taste in her mouth, a runny nose and a headache.
She also said her neck felt stiff and she her eyes were sensitive to light, according to a case report, published today in the journal JAMA.
The woman, in her forties, told medics she had recently been tested for Covid-19 ahead of an operation to repair a hernia.
But she said that shortly after the surgery she had developed a runny nose, headache and vomiting.
During an examination, medics at the University of Iowa Hospitals in Iowa City, US, discovered she had a mass in the middle of the right nasal cavity.
They drained the mass and it tested positive for a protein in cerebrospinal fluid, which is found in the brain or spine.
https://www.thesun.co.uk/news/12820053/coronavirus-swab-test-poked-womans-nose-brain-leak/
————————————-
Brain Scraper: Why Do Some COVID Tests Hurt So Much?
2021
https://www.webmd.com/lung/news/20210330/brain-scraper-why-do-some-covid-tests-hurt-so-much
————————————-
Goodbye, brain scrapers. COVID-19 tests now use gentler nose swabs
August 19, 2020
Early COVID-19 images of swabbing from Wuhan, China, looked more like an Ebola news story — health-care workers fully encased in personal protective equipment (PPE), inserting swabs so deeply that brain injury seemed imminent.
As COVID-19 (and testing) spread around the world, there were reports of “brain scraping”, “brain stabbing” or “brain tickling” swabs. Perhaps this was your experience early in the pandemic. Perhaps these stories have put you off getting tested so far.
But if you go to a drive-through clinic today, you’re likely to have a different swab, one that’s briefly inserted and not so far up as before.
So if fear of the swab itself is holding you back from getting tested, here’s what you need to know about these gentler swabs.
https://theconversation.com/goodbye-brain-scrapers-covid-19-tests-now-use-gentler-nose-swabs-144416
————————————-
Can Painful Complications Arise After A Covid-19 Nasal Swab Test?
2021
https://www.forbes.com/sites/anuradhavaranasi/2021/04/29/can-painful-complications-arise-after-a-covid-19-nasal-swab-test/?sh=4fd48ed92e49
————————————-
Saliva-Based COVID-19 Test Might Be Alternative to Deep Nasal Swab
April 27, 2020
https://www.webmd.com/lung/news/20200427/saliva-covid-test-alternative-to-deep-nasal-swab#1
————————————-
Says COVID-19 testing could be done with mouth swabs, so maybe deeper swabbing is “implanting something.”
July 7, 2020
https://www.politifact.com/factchecks/2020/jul/13/facebook-posts/no-proof-nasal-swabs-preferred-covid-test-are-impl/
————————————-
New rapid test uses magnetic nanoparticles to detect coronavirus antibodies
Feb 09, 2021
https://www.nanowerk.com/nanotechnology-news2/newsid=57247.php
————————————-
The Graphene-Based Sensor that Detects COVID in Less than 5 Minutes
May 10 2021
https://www.azosensors.com/article.aspx?ArticleID=2229
————————————-
Implanted microchip could be used to verify COVID-19 vax status
December 20th 2021
https://wwmt.com/news/coronavirus/implanted-microchip-could-be-used-to-verify-covid-19-vax-status
————————————-
“This is what Bill Gates and George Soros want to do… secretly stick you with a chip while testing you for the coronavirus… the Dems have a bill on the House floor ready to vote on it to require this.”
May 10, 2020
https://www.politifact.com/factchecks/2020/may/28/facebook-posts/theres-no-plot-microchip-people-during-covid-19-te/
————————————-
Morgellons disease fibers? Are COVID-19 nasal swabs really planting things in your brain?
Mar. 07, 2021
https://www.oregonlive.com/coronavirus/2021/03/morgellons-disease-fibers-are-covid-19-nasal-swabs-really-planting-things-in-your-brain-totally-false.html
————————————-
China assures its citizens they won’t waddle ‘like penguins’ following coronavirus anal swabs after ‘fake’ video goes viral
1 February 2021
China shut down a viral clip that shows people walking strangely in a hospital
Authorities denied that the video was filmed after people received anal swabs
A statement promised the virus-detecting method wouldn’t leave discomfort
Some experts believe such a test is more accurate than a nasal or throat swab
https://www.dailymail.co.uk/news/article-9168707/Coronavirus-China-blasts-fake-video-people-waddling-like-penguins-COVID-19-anal-swabs.html
————————————-
China Begins Invasive Coronavirus Anal Swab Testing on Public
February 1, 2021
The Chinese Communist Party (CCP) has begun anal swab testing for the novel coronavirus (SARS-CoV-2) in a questionable new approach to tackling an increasingly expanding COVID-19 pandemic.
On Jan. 20, Party mouthpiece CCTV reported the Beijing municipal government announced it had used anal swabbing to test 1,298 students, staff, and teachers at a school where a 9-year-old boy was allegedly diagnosed having contracted COVID-19 on Jan. 18.
CCTV quoted an infectious disease specialist from Beijing’s Youan Hospital as saying: “The asymptomatic carriers and COVID-19 patients with mild-symptoms can recover very quickly. In three to five days after the infection, their pharynx will be virus-free.” The specialist reasoned that: “Stool samples will contain the virus for a longer time … testing from the anus can improve the detection rate and reduce the false-negative rate.”
https://www.visiontimes.com/2021/02/01/the-chinese-communist-party-ccp-has-begun-collecting-anal-swab-samples-for-novel-coronavirus-sars-cov-2-testing-in-a-questionable-new-approach-to-tackling-an-increasingly-expanding-coronavirus-dis.html
————————————-
Explainer | Pandemic travel: tests for Covid-19, which ones China recognises, and are anal swabs necessary?
29 Nov, 2021
https://www.scmp.com/news/china/science/article/3157158/pandemic-travel-tests-covid-19-which-ones-china-recognises-and
————————————-
Are COVID-19 Tests Sterilized With a Chemical Linked to Cancer?
14 April 2021
https://www.snopes.com/fact-check/covid-19-tests-chemical-cancer/
Expected 2020 federal limitations on ethylene oxide emissions were paused in response to the pandemic.
What’s True
EtO is used in gaseous form to sterilize roughly half of all medical equipment manufactured in the U.S., including at least some nasal swabs used in diagnostic tests used to detect COVID-19. It is true that the chemical itself has been linked to cancer and other adverse health effects, specifically by exposure resulting from inhaling emissions from industrial facilities. However …
What’s False
Health experts told Snopes that it is false to say that COVID-19 tests sterilized with EtO can cause cancer. According to the U.S. Food and Drug Administration (FDA), medical equipment sterilized with EtO must meet recognized standards to ensure that levels of ethylene oxide on such equipment remain within safe limits. The FDA regards the use of EtO as a “safe and effective” method of sterilization.
————————————-
Ethylene Oxide
December 28, 2018
What is ethylene oxide?
At room temperature, ethylene oxide is a flammable colorless gas with a sweet odor. It is used primarily to produce other chemicals, including antifreeze. In smaller amounts, ethylene oxide is used as a pesticide and a sterilizing agent. The ability of ethylene oxide to damage DNA makes it an effective sterilizing agent but also accounts for its cancer-causing activity.
How are people exposed to ethylene oxide?
The primary routes of human exposure to ethylene oxide are inhalation and ingestion, which may occur through occupational, consumer, or environmental exposure. Because ethylene oxide is highly explosive and reactive, the equipment used for its processing generally consists of tightly closed and highly automated systems, which decreases the risk of occupational exposure.
Despite these precautions, workers and people who live near industrial facilities that produce or use ethylene oxide may be exposed to ethylene oxide through uncontrolled industrial emissions. The general population may also be exposed through tobacco smoke and the use of products that have been sterilized with ethylene oxide, such as medical products, cosmetics, and beekeeping equipment.
Which cancers are associated with exposure to ethylene oxide?
Lymphoma and leukemia are the cancers most frequently reported to be associated with occupational exposure to ethylene oxide. Stomach and breast cancers may also be associated with ethylene oxide exposure.
How can exposures be reduced?
The U.S. Occupational Safety and Health Administration has information about limiting occupational exposure to ethylene oxide.
https://www.cancer.gov/about-cancer/causes-prevention/risk/substances/ethylene-oxide
————————————-
Poison Control Center Issues Warning on Toxic Chemical in At-home COVID-19 Test Kits
Feb 27, 2022
https://news.ntd.com/poison-control-center-issues-warning-on-toxic-chemical-in-at-home-covid-19-test-kits_745487.html
Some at-home rapid COVID-19 tests contain a toxic chemical that may be harmful to both children and adults, according to health officials with the National Capital Poison Center.
“It is important to know that the extraction vial in many rapid antigen kits includes the chemical sodium azide as a preservative agent,” the Center said in a recent alert. “The BinaxNow, BD Veritor, Flowflex, and Celltrion DiaTrust COVID-19 rapid antigen kits all contain this chemical.”
Sodium azide is a colorless, odorless powder that testers dip cotton swabs into. The chemical is found in herbicides, pest control agents, and airbags for cars.
“Small doses of sodium azide can lower blood pressure, and larger doses may cause more serious health effects,” an advisory from Health Canada also said. “ProClin is also found in many kits. It contains chemicals that can cause skin and eye irritation, as well as allergic reactions.”
Some hospitals around the United States say they have received a surge in phone calls about exposures to the chemical.
“We started getting our first exposures to these test kits around early November,” said Sheila Goertemoeller, pharmacist and clinical toxicologist for the Cincinnati Children’s Hospital Medical Center. “It was, really, all ages.”
“Mostly, I’ve been very worried about our young children,” Goertemoeller said.
Accidental exposure is occurring among both children and adults, said Dr. Kelly Johnson-Arbor, with the National Capital Poison Center in Washington D.C., according to WNEP.
“People might mistake them for eye drops. Children might drop it onto their skin. Adults will sometimes mistakenly put them into their eyes,” said Johnson-Arbor.
“You don’t want to leave it on the skin because it could potentially cause an allergic reaction or a skin rash. If someone drinks the solution, it’s really important to contact poison control right away,” she added. “The solutions have different ingredients. Some have non-toxic ingredients and others have more dangerous ingredients.”
Officials told WNEP that there is no need to throw away the test kits, but people should be mindful when using them.
“Use them properly, dispose of them properly and it won’t cause an issue,” Dr. Jeffrey Jahre, with St. Luke’s University Health Network, told the outlet.
If you suspect you or someone you know has ingested the chemical, officials recommend not to make the person vomit. For eye exposures, rinse the eyes for 15 to 20 minutes with warm water. For skin exposures, rinse the skin well with tap water.
————————————-
Sodium azide
https://en.wikipedia.org/wiki/Sodium_azide
Sodium azide is the inorganic compound with the formula NaN3. It is used for the preparation of other azide compounds. It is an ionic substance, is highly soluble in water and is very acutely poisonous.
Biochemistry and biomedical uses
Sodium azide is a useful probe reagent and a preservative.
In hospitals and laboratories, it is a biocide; it is especially important in bulk reagents and stock solutions which may otherwise support bacterial growth where the sodium azide acts as a bacteriostatic by inhibiting cytochrome oxidase in gram-negative bacteria; however, some gram-positive bacteria (streptococci, pneumococci, lactobacilli) are intrinsically resistant.
Agricultural uses
It is used in agriculture for pest control of soil-borne pathogens such as Meloidogyne incognita or Helicotylenchus dihystera.
It is also used as a mutagen for crop selection of plants such as rice, barley or oats.
Safety considerations
Sodium azide has caused deaths for decades, and even minute amounts can cause symptoms. The toxicity of this compound is comparable to that of soluble alkali cyanides, although no toxicity has been reported from spent airbags.
It produces extrapyramidal symptoms with necrosis of the cerebral cortex, cerebellum, and basal ganglia. Toxicity may also include hypotension, blindness and hepatic necrosis. Sodium azide increases cyclic GMP levels in the brain and liver by activation of guanylate cyclase.
Sodium azide solutions react with metallic ions to precipitate metal azides, which can be shock sensitive and explosive. This should be considered for choosing a non-metallic transport container for sodium azide solutions in the laboratory. This can also create potentially dangerous situations if azide solutions should be directly disposed down the drain into a sanitary sewer system. Metal in the plumbing system could react, forming highly sensitive metal azide crystals which could accumulate over years. Adequate precautions are necessary for the safe and environmentally responsible disposal of azide solution residues.
————————————-
System and Method for Testing for COVID-19
Inventor: Richard A. ROTHSCHILD
Abstract
A method is provided for acquiring and transmitting biometric data (e.g., vital signs) of a user, where the data is analyzed to determine whether the user is suffering from a viral infection, such as COVID-19. The method includes using a pulse oximeter to acquire at least pulse and blood oxygen saturation percentage, which is transmitted wirelessly to a smartphone. To ensure that the data is accurate, an accelerometer within the smartphone is used to measure movement of the smartphone and/or the user. Once accurate data is acquired, it is uploaded to the cloud (or host), where the data is used (alone or together with other vital signs) to determine whether the user is suffering from (or likely to suffer from) a viral infection, such as COVID-19. Depending on the specific requirements, the data, changes thereto, and/or the determination can be used to alert medical staff and take corresponding actions.
https://patents.google.com/patent/US20200279585A1/en
————————————-
FACT CHECK: Was A Patent For A COVID-19 Test Issued In 2015? (Debated)
5:20 PM 08/10/2021
https://checkyourfact.com/2021/08/10/fact-check-patent-covid-19-test-issued-2015/
Fact Check:
The post includes a photo of what appears to be a patent application for “System and Method for Testing for COVID-19.” The patent’s abstract reads, “A method is provided for acquiring and transmitting biometric data (e.g., vital signs) of a user, where the data is analyzed to determine whether the user is suffering from a viral infection, such as COVID-19.” Text accompanying the image reads, “here is a patent from 2015……….yes it was all planned.”
While the priority date of the patent is Oct. 13, 2015, the actual patent shown in the Facebook image is not from then. The priority date is the earliest date of a filing in a family of patent applications, not the date a specific patent was published, patent attorney Vic Lin wrote in the Patent Trademark Blog.
“If an applicant has filed a number of related patent applications, the priority date would be the filing date of the earliest patent filing that first disclosed the invention,” Lin wrote. (RELATED: Did Bill Gates Patent A ‘CV19-N95’ Face Mask Design Years Before The COVID-19 Pandemic)
The patent for “System and Method for Testing COVID-19” was filed May 17, 2020, and published on Sept. 3, 2020, according to the patent summary published in the National Library of Medicine. The patent application on the U.S. Patent and Trademark Office’s (USPTO) website shows other related patent documents, with the earliest one being filed Oct. 13, 2015.
The Related U.S. Application Data section of the 2020 patent application suggests it is a continuation-in-part application. This is a type of patent application that adds on to claims or inventions described in previous applications, according to USPTO.
Check Your Fact found that the related patent documents in the family that includes the “System and Method for Testing COVID-19” patent do not mention COVID-19. For example, one application was filed on April 24, 2017, and published in August 2017, and again in March 2019. At that time, it was titled, “System and Method for Using, Processing, and Displaying Biometric Data,” and did not mention COVID-19 or the coronavirus at any point.
Patents filed in February 2019 and December 2019 were also named “System and Method for Using, Processing, and Displaying Biometric Data,” and did not mention COVID-19. The Oct. 13, 2015, and Oct. 13, 2016 patent applications could not be accessed.
Furthermore, COVID-19 first emerged in Wuhan, China, in late 2019, according to the World Health Organization, meaning it would be impossible for the original patent application from 2015 to be related to the virus.
————————————-
Proof That Rothschilds Patented Covid-19 Biometric Tests In 2015 And 2017
Jul 08, 2021
https://www.oom2.com/t76111-proof-that-rothschilds-patented-covid-19-biometric-tests-in-2015-and-2017
Ultimate Proof: Covid-19 Was Planned to Usher in the New World Order: We Have Proof That Rothschilds Patented Covid-19 Biometric Tests In 2015 And 2017
Proof That Rothschilds Patented Covid-19 Biometric Tests In 2015 And 2017 5bd79c63a310eff369034103
Jack Metir Uncategorized July 6, 2021
It’s not disputable, since the information comes from official patent registries in the Netherlands and US. And we have all the documentation.
As we’ve shown in previous exposes, the whole Covidiocracy is a masquerade and a simulation long prepared by The World Bank / IMF / The Rothschilds / World Economic Forum (basically the world’s “elite”, the 0.1%) and their lemmings, with Rockefeller partnership.
Our newest discoveries further these previous revelations.
FIRST REGISTRATION: NETHERLANDS, 2015
Source: Dutch Government patent registry website
A method is provided for acquiring and transmitting biometric data (e.g., vital signs) of a user, where the data is analyzed to determine whether the user is suffering from a viral infection, such as COVID-19.
The method includes using a pulse oximeter to acquire at least pulse and blood oxygen saturation percentage, which is transmitted wirelessly to a smartphone.
To ensure that the data is accurate, an accelerometer within the smartphone is used to measure movement of the smartphone and/or the user.
Once accurate data is acquired, it is uploaded to the cloud (or host), where the data is used (alone or together with other vital signs) to determine whether the user is suffering from (or likely to suffer from) a viral infection, such as COVID-19.
Depending on the specific requirements, the data, changes thereto, and/or the determination can be used to alert medical staff and take corresponding actions.
————————————-
Ultimate Proof: Covid-19 Was Planned to Usher in the New World Order: We Have Proof That Rothschilds Patented Covid-19 Biometric Tests In 2015 And 2017
July 6, 2021
https://theblogginghounds.com/2021/07/07/ultimate-proof-covid-19-was-planned-to-usher-in-the-new-world-order-we-have-proof-that-rothschilds-patented-covid-19-biometric-tests-in-2015-and-2017/
————————————-
System and Method for Testing for COVID-19
Patent US-2020279585-A1
Inventor
ROTHSCHILD RICHARD A (GB)
Assignee
ROTHSCHILD RICHARD A (GB)
Date
Priority
2015/10/13
Abstract
New Window
A method is provided for acquiring and transmitting biometric data (e.g., vital signs) of a user, where the data is analyzed to determine whether the user is suffering from a viral infection, such as COVID-19. The method includes using a pulse oximeter to acquire at least pulse and blood oxygen saturation percentage, which is transmitted wirelessly to a smartphone. To ensure that the data is accurate, an accelerometer within the smartphone is used to measure movement of the smartphone and/or the user. Once accurate data is acquired, it is uploaded to the cloud (or host), where the data is used (alone or together with other vital signs) to determine whether the user is suffering from (or likely to suffer from) a viral infection, such as COVID-19. Depending on the specific requirements, the data, changes thereto, and/or the determination can be used to alert medical staff and take corresponding actions.
2015/10/13
Source: Google Patents
URL: https://patents.google.com/patent/US20200279585A1
Description: System and Method for Testing for COVID-19
License Note: “Google Patents Research Data” by Google, based on data provided by IFI CLAIMS Patent Services and OntoChem, is licensed under a Creative Commons Attribution 4.0 International License.
License URL: https://creativecommons.org/licenses/by/4.0/legalcode
3.2 Filing Date
2020/05/17
Source: Google Patents
URL: https://patents.google.com/patent/US20200279585A1
Description: System and Method for Testing for COVID-19
License Note: “Google Patents Research Data” by Google, based on data provided by IFI CLAIMS Patent Services and OntoChem, is licensed under a Creative Commons Attribution 4.0 International License.
License URL: https://creativecommons.org/licenses/by/4.0/legalcode
3.3Publication Date
2020/09/03
Source: Google Patents
URL: https://patents.google.com/patent/US20200279585A1
Description: System and Method for Testing for COVID-19
License Note: “Google Patents Research Data” by Google, based on data provided by IFI CLAIMS Patent Services and OntoChem, is licensed under a Creative Commons Attribution 4.0 International License.
License URL: https://creativecommons.org/licenses/by/4.0/legalcode
https://pubchem.ncbi.nlm.nih.gov/patent/US-2020279585-A1
————————————-
Tulane Researchers Design Nanotechnology Blood Test to Find Hidden COVID-19 Infections
2021-08-12
Tulane University researchers have developed a new type of blood test to find these hidden infections using nanoparticles to detect fragments of the virus released by infected cells anywhere in the body. Because the test uses a screening target that remains stable in the blood, it can detect COVID-19 weeks after initial infection, according to a new study published in the journal Nature Nanotechnology.
The test analyzes small lipid-enclosed bubbles of cell material called extracellular vesicles (EVs). These vesicles accumulate in the blood and protect their contents from being destroyed by enzymes. Cells infected by SARS-CoV-2 secrete EVs that contain RNA from the virus. Researchers captured these EVs using an antibody and then fused them with synthetic lipid vesicles loaded with a testing reagent. The blood test uses reverse transcription PCR to amplify the RNA target region and CRISPR to amplify the signal produced by this target to detect an infection.
https://statnano.com/news/69563/Tulane-Researchers-Design-Nanotechnology-Blood-Test-to-Find-Hidden-COVID-19-Infections
————————————-
Insights from nanotechnology in COVID-19: prevention, detection, therapy and immunomodulation
2021 May 17
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8127834/
————————————-
Electric field-driven microfluidics for rapid CRISPR-based diagnostics and its application to detection of SARS-CoV-2
2020
https://www.pnas.org/content/117/47/29518
————————————-
New CRISPR-based diagnostic test detects COVID-19 variants from saliva samples
Aug 6 2021
https://www.news-medical.net/news/20210806/New-CRISPR-based-diagnostic-test-detects-COVID-19-variants-from-saliva-samples.aspx
————————————-
An engineered CRISPR-Cas12a variant and DNA-RNA hybrid guides enable robust and rapid COVID-19 testing
19 March 2021
————————————-
New kind of CRISPR technology to target RNA, including RNA viruses like coronavirus
March 16, 2020
https://www.sciencedaily.com/releases/2020/03/200316141514.htm
————————————–
Could Crispr Be Humanity’s Next Virus Killer?
03.18.2020
https://www.wired.com/story/could-crispr-be-the-next-virus-killer/
————————————-
CRISPR-Based Anti-Viral Therapy Could One Day Foil the Flu—and COVID-19
March 16th, 2021
https://directorsblog.nih.gov/2021/03/16/crispr-based-anti-viral-therapy-could-one-day-foil-the-flu-and-covid-19/
————————————-
New research suggests CRISPR can destroy virus that causes COVID-19
July 30, 2021
https://allianceforscience.cornell.edu/blog/2021/07/new-research-suggests-crispr-can-destroy-virus-that-causes-covid-19/
————————————-
CRISPRclean Technology – Introduction
Oct 18, 2021
https://www.youtube.com/watch?v=Rv3vRHA10gs
————————————-
Double-Barreled CRISPR Technology as a Novel Treatment Strategy For COVID-19
2020 Aug 27
https://www.ncbi.nlm.nih.gov/labs/pmc/articles/PMC7469881/
————————————-
CRISPR-Cas9 genome editing using targeted lipid nanoparticles for cancer therapy
18 Nov 2020
https://advances.sciencemag.org/content/6/47/eabc9450.full
————————————-
How does the mRNA/lipid nanoparticle platform promote protective adaptive immunity against SARS-CoV-2?
Aug 5 2021
https://www.news-medical.net/news/20210805/How-does-the-mRNAlipid-nanoparticle-platform-promote-protective-adaptive-immunity-against-SARS-CoV-2.aspx
————————————-
Fact check: Lipid nanoparticles in a COVID-19 vaccine are there to transport RNA molecules
December 5, 2020
https://www.reuters.com/article/uk-factcheck-vaccine-nanoparticles-idUSKBN28F0I9
————————————-
New genetic method of using CRISPR to eliminate COVID-19 virus genomes in cells
March 2020
https://bioengineering.stanford.edu/research-impact/prevent-covid-19/new-genetic-method-using-crispr-eliminate-covid-19-virus-genomes
————————————-
CRISPR Breakthrough Blocks SARS-CoV-2 Virus Replication in Early Lab Tests
14 JULY 2021
https://www.sciencealert.com/cell-study-suggests-we-might-be-able-to-stop-sars-cov-2-from-replicating
————————————-
COVID-19 one year later: a retrospect of CRISPR-Cas system in combating COVID-19
2021
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8193275/
————————————-
| Synthetic recombinant bat SARS-like coronavirus is infectious in cultured cells and in mice December 16, 2008 https://www.pnas.org/content/105/50/19944 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ———————————- | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| COVID-19: Synthetic SARS-CoV-2 could be used as antiviral therapy July 14, 2021 Researchers created a synthetic virus that can prevent the growth of the real virus https://www.theweek.in/news/sci-tech/2021/07/14/covid-19–synthetic-sars-cov-2-could-be-used-as-antiviral-therap.html | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ——————————– | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Synthetic Antibody Prevents COVID Virus Replication February 3rd, 2021 https://www.medgadget.com/2021/02/synthetic-antibody-prevents-covid-virus-replication.html | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ——————————— | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Can we fight SARS-CoV-2 with SARS-CoV-2? July 12, 2021 https://www.medicalnewstoday.com/articles/can-we-fight-sars-cov-2-with-sars-cov-2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Coronaviridae
https://en.wikipedia.org/wiki/Coronaviridae
Coronaviridae is a family of enveloped, positive-strand RNA viruses which infect amphibians, birds, and mammals. The group includes the subfamilies Letovirinae and Orthocoronavirinae; the members of the latter are known as coronaviruses.
The viral genome is 26–32 kilobases in length. The particles are typically decorated with large (~20 nm), club- or petal-shaped surface projections (the “peplomers” or “spikes”), which in electron micrographs of spherical particles create an image reminiscent of the solar corona.
Orthocoronavirinae taxonomy
Orthocoronavirinae
Alphacoronavirus
Colacovirus
Bat coronavirus CDPHE15
Decacovirus
Bat coronavirus HKU10
Rhinolophus ferrumequinum alphacoronavirus HuB-2013
Duvinacovirus
Human coronavirus 229E
Luchacovirus
Lucheng Rn rat coronavirus
Minacovirus
Mink coronavirus 1
Minunacovirus
Miniopterus bat coronavirus 1
Miniopterus bat coronavirus HKU8
Myotacovirus
Myotis ricketti alphacoronavirus Sax-2011
Nyctacovirus
Nyctalus velutinus alphacoronavirus SC-2013
Pipistrellus kuhlii coronavirus 3398
Pedacovirus
Porcine epidemic diarrhea virus
Scotophilus bat coronavirus 512
Rhinacovirus
Rhinolophus bat coronavirus HKU2
Setracovirus
Human coronavirus NL63
NL63-related bat coronavirus strain BtKYNL63-9b
Soracovirus
Sorex araneus coronavirus T14
Sunacovirus
Suncus murinus coronavirus X74
Tegacovirus
Alphacoronavirus 1
Betacoronavirus
Embecovirus
Betacoronavirus 1
Human coronavirus OC43
China Rattus coronavirus HKU24
Human coronavirus HKU1
Murine coronavirus
Myodes coronavirus 2JL14
Hibecovirus
Bat Hp-betacoronavirus Zhejiang2013
Merbecovirus
Hedgehog coronavirus 1
Middle East respiratory syndrome-related coronavirus (MERS-CoV)
Pipistrellus bat coronavirus HKU5
Tylonycteris bat coronavirus HKU4
Nobecovirus
Eidolon bat coronavirus C704
Rousettus bat coronavirus GCCDC1
Rousettus bat coronavirus HKU9
Sarbecovirus
Severe acute respiratory syndrome–related coronavirus
Severe acute respiratory syndrome coronavirus (SARS-CoV)
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2, cause of COVID-19)
Gammacoronavirus
Brangacovirus
Goose coronavirus CB17
Cegacovirus
Beluga whale coronavirus SW1
Igacovirus
Avian coronavirus
Avian coronavirus 9203
Duck coronavirus 2714
Deltacoronavirus
Andecovirus
Wigeon coronavirus HKU20
Buldecovirus
Bulbul coronavirus HKU11
Common moorhen coronavirus HKU21
Coronavirus HKU15
Munia coronavirus HKU13
White-eye coronavirus HKU16
Herdecovirus
Night heron coronavirus HKU19
————————————————————————–
Bat Virome
https://en.wikipedia.org/wiki/Bat_virome
Contents
1 Viral diversity
1.1 Transmission to humans
1.2 Bats compared to other viral reservoirs
1.3 Sampling
2 Double-stranded DNA viruses
2.1 Adenoviruses
2.2 Herpesviruses
2.3 Papillomaviruses
3 Single-stranded DNA viruses
3.1 Anelloviruses
3.2 Circoviruses
3.3 Parvoviruses
4 Double-stranded RNA viruses
4.1 Reoviruses
4.1.1 Zoonotic
4.1.2 Other
5 Positive-sense single-stranded RNA viruses
5.1 Astroviruses
5.2 Caliciviruses
5.3 Coronaviruses
5.3.1 SARS-CoV, SARS-CoV-2, and MERS-CoV
5.3.2 Other
5.4 Flaviviruses
5.5 Picornaviruses
6 Negative-sense single-stranded RNA viruses
6.1 Arenaviruses
6.2 Hantaviruses
6.3 Filoviruses
6.3.1 Marburgvirus and Ebolavirus
6.3.2 Other
6.4 Rhabdoviruses
6.4.1 Rabies-causing viruses
6.4.2 Other
6.5 Orthomyxoviruses
6.6 Paramyxoviruses
6.6.1 Hendra, Nipah, and Menangle viruses
6.6.2 Other
6.7 Togaviruses
7 Positive-sense single-stranded RNA viruses that replicate through a DNA intermediate
7.1 Retroviruses
8 Double-stranded DNA viruses that replicate through a single-stranded RNA intermediate
8.1 Hepadnaviruses
9 See also
10 References
————————————————————————–
Rc-o319
https://en.wikipedia.org/wiki/Rc-o319
Rc-o319 is a bat-derived strain of severe acute respiratory syndrome–related coronavirus collected in Little Japanese horseshoe bats (Rhinolophus cornutus) from sites in Iwate, Japan.[1] Its has 81% similarity to SARS-CoV-2 and is the earliest strain branch of the SARS-CoV-2 related coronavirus.
A phylogenetic tree based on whole-genome sequences of SARS-CoV-2 and related coronaviruses is:
————————————————————————–
RpYN06
https://en.wikipedia.org/wiki/RpYN06
Bat coronavirus RpYN06 is a SARS-like betacoronavirus that infects the horseshoe bat Rhinolophus pusillus, it is the second closest known relative of SARS-CoV-2 with a 94.48% sequence identity.
————————————————————————–
RmYN02
https://en.wikipedia.org/wiki/RmYN02
RmYN02 is a bat-derived strain of Severe acute respiratory syndrome–related coronavirus. It was discovered in bat droppings collected between May and October 2019 from sites in Mengla County, Yunnan Province, China. It is the second-closest known relative of SARS-CoV-2, the virus strain that causes COVID-19, sharing 93.3% nucleotide identity at the scale of the complete virus genome. RmYN02 contains an insertion at the S1/S2 cleavage site in the spike protein, similar to SARS-CoV-2, suggesting that such insertion events can occur naturally, which was the subject of a paper sent to Nature.
————————————————————————–
RaTG13
https://en.wikipedia.org/wiki/RaTG13
Bat coronavirus RaTG13 is a SARS-like betacoronavirus that infects the horseshoe bat Rhinolophus affinis. It was discovered in 2013 in bat droppings from a mining cave near the town of Tongguan in Mojiang county in Yunnan, China. As of 2020, it is the closest known relative of SARS-CoV-2, the virus that causes COVID-19.
————————————————————————–
COVID-19 Virus Binds to Human Cells 1,000 Times Tighter Than its Closest Relative
7-10-20
https://www.newsweek.com/sars-cov-2-coronavirus-infect-cells-1516882
————————————————————————–
Peptide Scanning of SARS-CoV and SARS-CoV-2 Spike Protein Subunit 1
Reveals Potential Additional Receptor Binding Sites
2021
https://www.biorxiv.org/content/10.1101/2021.08.16.456470v1.full.pdf
————————————————————————–
All about ACE-2 — the molecule that helps novel coronavirus invade cells
13 May 2020
SARS-CoV-2 has a high-binding capacity for ACE2 — between 10 and 20 times more that of original SARS virus
https://www.downtoearth.org.in/news/science-technology/all-about-ace-2-the-molecule-that-helps-novel-coronavirus-invade-cells-71089
————————————————————————–
SARS-CoV-2 needs cholesterol to invade cells and form mega cells
January 22, 2021
https://phys.org/news/2021-01-sars-cov-cholesterol-invade-cells-mega.html
————————————————————————–
Viroid
https://en.wikipedia.org/wiki/Viroid
Viroids are small single-stranded, circular RNAs that are infectious pathogens. Unlike viruses, they have no protein coating. All known viroids are inhabitants of angiosperms (flowering plants), and most cause diseases, whose respective economic importance to humans varies widely.
The first discoveries of viroids in the 1970s triggered the historically third major extension of the biosphere—to include smaller lifelike entities —after the discoveries in 1675 by Antonie van Leeuwenhoek (of the “subvisible” microorganisms) and in 1892–1898 by Dmitri Iosifovich Ivanovsky and Martinus Beijerinck (of the “submicroscopic” viruses). The unique properties of viroids have been recognized by the International Committee on Taxonomy of Viruses, in creating a new order of subviral agents.
The first recognized viroid, the pathogenic agent of the potato spindle tuber disease, was discovered, initially molecularly characterized, and named by Theodor Otto Diener, plant pathologist at the U.S Department of Agriculture’s Research Center in Beltsville, Maryland, in 1971. This viroid is now called potato spindle tuber viroid, abbreviated PSTVd. The Citrus exocortis viroid (CEVd) was discovered soon thereafter, and together understanding of PSTVd and CEVd shaped the concept of the viroid.
Although viroids are composed of nucleic acid, they do not code for any protein. The viroid’s replication mechanism uses RNA polymerase II, a host cell enzyme normally associated with synthesis of messenger RNA from DNA, which instead catalyzes “rolling circle” synthesis of new RNA using the viroid’s RNA as a template. Viroids are often ribozymes, having catalytic properties that allow self-cleavage and ligation of unit-size genomes from larger replication intermediates.
Diener initially hypothesized in 1989 that viroids may represent “living relics” from the widely assumed, ancient, and non-cellular RNA world, and others have followed this conjecture. Following the discovery of retrozymes, it is now thought that viroids and other viroid-like elements may derive from this newly found class of retrotransposon.
The human pathogen hepatitis D virus is a subviral agent similar in structure to a viroid.
————————————————————————–
List of virus species
https://en.wikipedia.org/wiki/List_of_virus_species
This is a list of all virus species, including satellites and viroids. Excluded are other ranks, and other non-cellular life such as prions. Also excluded are common names and obsolete names for viruses.
For a list of virus genera, see List of virus genera.
For a list of virus families and subfamilies, see List of virus families and subfamilies.
For a list of virus realms, subrealms, kingdoms, subkingdoms, phyla, subphyla, classes, subclasses, orders, and suborders, see List of higher virus taxa.
————————————————————————–
Astrovirus
https://en.wikipedia.org/wiki/Astrovirus
Astroviruses are a type of virus that was first discovered in 1975 using electron microscopes following an outbreak of diarrhea in humans. In addition to humans, astroviruses have now been isolated from numerous mammalian animal species (and are classified as genus Mammoastrovirus) and from avian species such as ducks, chickens, and turkey poults (classified as genus Avastrovirus). Astroviruses are 28–35 nm diameter, icosahedral viruses that have a characteristic five- or sixpointed star-like surface structure when viewed by electron microscopy. Along with the Picornaviridae and the Caliciviridae, the Astroviridae comprise a third family of nonenveloped viruses whose genome is composed of plus-sense, single-stranded RNA. Astrovirus has a non-segmented, single stranded, positive sense RNA genome within a non-enveloped icosahedral capsid. Human astroviruses have been shown in numerous studies to be an important cause of gastroenteritis in young children worldwide. In animals, Astroviruses also cause infection of the gastrointestinal tract but may also result in encephalitis (humans and cattle), hepatitis (avian) and nephritis (avian).
————————————————————————–
Betaretrovirus
Betaretrovirus is a genus of the Retroviridae family. It has type B or type D morphology. The type B is common for a few exogenous, vertically transmitted and endogenous viruses of mice; some primate and sheep viruses are the type D.
Examples are Mouse mammary tumor virus, enzootic nasal tumor virus (ENTV-1, ENTV-2), and simian retrovirus types 1, 2 and 3 (SRV-1, SRV-2, SRV-3).
https://en.wikipedia.org/wiki/Betaretrovirus
————————————————————————–
Researchers wind up a 40 year old debate on betaretrovirus infection in humans
February 19, 2015
https://medicalxpress.com/news/2015-02-year-debate-betaretrovirus-infection-humans.html
————————————————————————–
Caliciviridae
The Caliciviridae are a family of “small round structured” viruses, members of Class IV of the Baltimore scheme. Caliciviridae bear resemblance to enlarged picornavirus and was formerly a separate genus within the picornaviridae. They are positive-sense, single-stranded RNA which is not segmented. Thirteen species are placed in this family, divided among eleven genera. Diseases associated with this family include feline calicivirus (respiratory disease), rabbit hemorrhagic disease virus (often-fatal hemorrhages), and Norwalk group of viruses (gastroenteritis). Caliciviruses naturally infect vertebrates, and have been found in a number of organisms such as humans, cattle, pigs, cats, chickens, reptiles, dolphins and amphibians. The caliciviruses have a simple construction and are not enveloped. The capsid appears hexagonal/spherical and has icosahedral symmetry (T=1 or T=3) with a diameter of 35–39 nm.
Caliciviruses are not very well studied because until recently, they could not be grown in culture, and they have a very narrow host range and no suitable animal model. However, the recent application of modern genomic technologies has led to an increased understanding of the virus family.[5] A recent isolate from rhesus monkeys—Tulane virus—can be grown in culture, and this system promises to increase understanding of these viruses.
https://en.wikipedia.org/wiki/Caliciviridae
————————————————————————–
Decacovirus
Decacovirus is a subgenus of viruses in the genus Alphacoronavirus.
https://en.wikipedia.org/wiki/Decacovirus
————————————————————————–
Enterovirus
https://en.wikipedia.org/wiki/Enterovirus
Enterovirus is a genus of positive-sense single-stranded RNA viruses associated with several human and mammalian diseases. Enteroviruses are named by their transmission-route through the intestine (enteric meaning intestinal).
Serologic studies have distinguished 71 human enterovirus serotypes on the basis of antibody neutralization tests. Additional antigenic variants have been defined within several of the serotypes on the basis of reduced or nonreciprocal cross-neutralization between variant strains. On the basis of their pathogenesis in humans and animals, the enteroviruses were originally classified into four groups, polioviruses, Coxsackie A viruses (CA), Coxsackie B viruses (CB), and echoviruses, but it was quickly realized that there were significant overlaps in the biological properties of viruses in the different groups. Enteroviruses isolated more recently are named with a system of consecutive numbers: EV-D68, EV-B69, EV-D70, EV-A71, etc, where genotyping is based on the VP1 capsid region.
Enteroviruses affect millions of people worldwide each year and are often found in the respiratory secretions (e.g., saliva, sputum, or nasal mucus) and stool of an infected person. Historically, poliomyelitis was the most significant disease caused by an enterovirus, namely poliovirus. There are 81 non-polio and 3 polio enteroviruses that can cause disease in humans. Of the 81 non-polio types, there are 22 Coxsackie A viruses, 6 Coxsackie B viruses, 28 echoviruses, and 25 other enteroviruses.
Poliovirus, as well as coxsackie and echovirus, is spread through the fecal-oral route. Infection can result in a wide variety of symptoms, including those of: mild respiratory illness (the common cold), hand, foot and mouth disease, acute hemorrhagic conjunctivitis, aseptic meningitis, myocarditis, severe neonatal sepsis-like disease, acute flaccid paralysis, and the related acute flaccid myelitis.
Contents
1 Virology
2 Member viruses
2.1 Enterovirus A – L
2.2 Non-cytolytic (non-cytopathic) enterovirus
2.3 Enterovirus D68
2.4 Enterovirus A71
2.5 Poliovirus
3 Diseases caused by enterovirus infection
3.1 Suspected diseases
3.2 Possible correlations being studied
4 Symptoms
5 Treatment
6 Taxonomy
————————————————————————–
Gammaretrovirus
Gammaretroviruses have a genus-specific PXXIGPNPVLADQ motif at the end of their RBD that apparently interacts with the SPHQ motif and influences fusion.
https://www.sciencedirect.com/topics/biochemistry-genetics-and-molecular-biology/gammaretrovirus
————————————————————————–
Hepadnaviridae
Hepadnaviridae[a] is a family of viruses. Humans, apes, and birds serve as natural hosts. There are currently 18 species in this family, divided among 5 genera. Its best-known member is hepatitis B virus. Diseases associated with this family include: liver infections, such as hepatitis, hepatocellular carcinomas (chronic infections), and cirrhosis. It is the sole family in the order Blubervirales
https://en.wikipedia.org/wiki/Hepadnaviridae
————————————————————————–
Letovirinae
https://en.wikipedia.org/wiki/Letovirinae
Letovirinae is a subfamily of viruses within the family Coronaviridae, where it is the only subfamily besides the more diverse Orthocoronavirinae (coronaviruses). Letovirinae contains one accepted genus, Alphaletovirus, which contains one accepted subgenus, Milecovirus, which contains one accepted species, Microhyla letovirus 1 (MLeV). This species was discovered in 2018 and is hosted by the ornate chorus frog (Microhyla fissipes).
Other, as yet unaccepted species in the Letovirinae have been discovered in Pacific salmon (Oncorhynchus), and in Murray River carp (Cyprinus).
————————————————————————–
Endangered wild salmon infected by newly discovered viruses
2019
https://elifesciences.org/articles/47615.pdf
————————————————————————–
Lloviu virus
The species Lloviu cuevavirus is the taxonomic home of a virus that forms filamentous virion, Lloviu virus (LLOV). The species is included in the genus Cuevavirus. LLOV is a distant relative of the commonly known Ebola virus and Marburg virus.
https://en.wikipedia.org/wiki/Lloviu_virus
————————————————————————–
Oncolytic virus
https://en.wikipedia.org/wiki/Oncolytic_virus
An oncolytic virus is a virus that preferentially infects and kills cancer cells. As the infected cancer cells are destroyed by oncolysis, they release new infectious virus particles or virions to help destroy the remaining tumour. Oncolytic viruses are thought not only to cause direct destruction of the tumour cells, but also to stimulate host anti-tumour immune system responses.
The potential of viruses as anti-cancer agents was first realised in the early twentieth century, although coordinated research efforts did not begin until the 1960s. A number of viruses including adenovirus, reovirus, measles, herpes simplex, Newcastle disease virus, and vaccinia have been clinically tested as oncolytic agents. Most current oncolytic viruses are engineered for tumour selectivity, although there are naturally occurring examples such as reovirus and the senecavirus, resulting in clinical trials.
The first oncolytic virus to be approved by a national regulatory agency was genetically unmodified ECHO-7 strain enterovirus RIGVIR, which was approved in Latvia in 2004 for the treatment of skin melanoma; the approval was withdrawn in 2019. An oncolytic adenovirus, a genetically modified adenovirus named H101, was approved in China in 2005 for the treatment of head and neck cancer. In 2015, talimogene laherparepvec (OncoVex, T-VEC), an oncolytic herpes virus which is a modified herpes simplex virus, became the first oncolytic virus to be approved for use in the U.S. and European Union, for the treatment of advanced inoperable melanoma.
————————————————————————–
Spumaretrovirinae
https://en.wikipedia.org/wiki/Spumaretrovirinae
Spumaretrovirinae, commonly called spumaviruses (spuma, Latin for “foam”) or foamyviruses, is a subfamily of the Retroviridae family. Spumaviruses are exogenous viruses that have specific morphology with prominent surface spikes. The virions contain significant amounts of double-stranded full-length DNA, and assembly is rather unusual in these viruses. Spumaviruses are unlike most enveloped viruses in that the envelope membrane is acquired by budding through the endoplasmic reticulum instead of the cytoplasmic membrane. Some spumaviruses, including the equine foamy virus (EFV), bud from the cytoplasmic membrane.
Some examples of these viruses are simian foamy virus and the human foamy virus.
While spumaviruses will form characteristic large vacuoles in their host cells while in vitro, there is no disease association in vivo.
————————————————————————–
Retrovirus
https://en.wikipedia.org/wiki/Retrovirus
————————————————————————–
What is the Difference Between Adenovirus and Retrovirus
July 2, 2019
The main difference between adenovirus and retrovirus is that adenovirus is the largest, non-enveloped virus, whereas retrovirus is an enveloped virus. Furthermore, the genome of adenovirus is double-stranded DNA, while the genome of a retrovirus is single-stranded RNA (+). Moreover, adenovirus infects both dividing and non-dividing cells, and it causes upper respiratory tract infections in both adults and children, while the retrovirus infects only dividing cells, and it has a higher tendency to cause diseases.
Adenovirus and retrovirus are two infectious agents that come into contact with living cells to undergo replication. Moreover, they can be used as vectors to deliver desired DNA fragments to the host cell during gene therapy.
https://pediaa.com/what-is-the-difference-between-adenovirus-and-retrovirus/
————————————————————————–
Host-virus Interactions: Closing the net on retroviruses
Jul 7, 2016
https://elifesciences.org/articles/18243
————————————————————————–
Reston virus
Reston virus (RESTV) is one of six known viruses within the genus Ebolavirus. Reston virus causes Ebola virus disease in non-human primates; unlike the other five ebolaviruses, it is not known to cause disease in humans, but has caused asymptomatic infections. Reston virus was first described in 1990 as a new “strain” of Ebola virus (EBOV). It is the single member of the species Reston ebolavirus, which is included into the genus Ebolavirus, family Filoviridae, order Mononegavirales. Reston virus is named after Reston, Virginia, US, where the virus was first discovered.
https://en.wikipedia.org/wiki/Reston_virus
————————————————————————–
Xenotropic murine leukemia virus–related virus
https://en.wikipedia.org/wiki/Xenotropic_murine_leukemia_virus%E2%80%93related_virus
Xenotropic murine leukemia virus–related virus (XMRV) is a retrovirus which was first described in 2006 as an apparently novel human pathogen found in tissue samples from men with prostate cancer. Initial reports erroneously linked the virus to prostate cancer and later to chronic fatigue syndrome (CFS), leading to considerable interest in the scientific and patient communities, investigation of XMRV as a potential cause of multiple medical conditions, and public-health concerns about the safety of the donated blood supply.
Xenotropic viruses replicate or reproduce in cells other than those of the host species. Murine refers to the rodent family Muridae, which includes common household rats and mice.
Subsequent research established that XMRV was in fact a laboratory contaminant, rather than a novel pathogen. XMRV was generated unintentionally in the laboratory, through genetic recombination between two mouse retroviruses during propagation of a prostate-cancer cell line in the mid-1990s. False-positive detection of XMRV may also occur because of contamination of clinical specimens and laboratory reagents with other mouse retroviruses or related nucleic acids. Most scientific publications claiming an association of XMRV with CFS or prostate cancer have been retracted, and allegations of research misconduct were leveled against at least one CFS investigator. There is no evidence that XMRV can infect humans, nor that XMRV is associated with or causes any human disease.
————————————————————————–
Xenotropic Virus
a retrovirus that does not produce disease in its natural host and replicates only in tissue culture cells derived from a different species.
https://medical-dictionary.thefreedictionary.com/xenotropic+virus
————————————————————————–
Ortervirales: New Virus Order Unifying Five Families of Reverse-Transcribing Viruses.
2018
https://europepmc.org/article/MED/29618642
————————————————————————–
Top 10 Mysterious Viruses
October 14, 2016
https://listverse.com/2016/10/14/top-10-mysterious-viruses/
#10: Black Widow Virus
Scientists have recently discovered a virus containing the gene for black widow poison.
The WO virus specifically targets Wolbachia bacteria in arthropods. Latrotoxin kills by poking holes in cell membranes. It is believed that the venom genes allow the WO virus to break through cells and evade the host’s immune system. This is the first time animal genes have been seen in bacteriophages—bacteria-targeting viruses.
Experts hypothesize that the virus picked up the genetic material after breaking out of a Wolbachia bacterium into a black widow cell. However, it is possible the spider stole the gene from the WO virus.
#9: Infertility Virus
A mysterious viral infection may be the cause of half of unexplained cases of infertility. In one-quarter of infertility cases—roughly one in 70 women under the age of 44—doctors cannot find a cause. An Italian research team discovered that a virus in the herpes family is to blame. It causes immune reactions that make the womb inhospitable for an embryo. Customized anti-viral treatment could offer help.
The team studied 30 mothers and 30 women with unexplained infertility. 13 of the infertile women were infected with HHV-6A. None of the mothers had it. This herpes variety was discovered over 30 years ago but remains a mystery. HHV-6A infection releases estradiol hormone, which triggers ovulation and prepares a womb for fertilization.
#8: Survivor Virus
Scientists have recently unlocked the secrets to a virus that can survive boiling acid.
The SIRV2 virus infects a microbe called Sulfolobus islandicus, which lives in acidic hot springs where temperatures top 80 degrees Celsius (175 °F). Using a Titan Krios electron microscope to examine the specimens in previously unimaginable detail, scientists have unlocked the basic mechanism of resistance to heat, desiccation, and ultra-violet radiation.
SIRV2 forces genetic material into a protective structural state called A-formation to survive extreme conditions. The mechanism is remarkably similar to the spores bacteria form to survive such environments. These spores are known to cause hard to combat diseases like anthrax. Scientists plan on using these survival mechanisms to design a DNA package for gene therapy.
#7: Multicomponent Virus
Normal viruses have all their genes in one viral particle. This viral ball attaches to a cell, opens, and injects its genetic material inside. The host cell begins replicating the virus. Once enough copies are made, they kill the cell, break free, and infect more.
The Guaico Culex virus is different. To become infected, a cell needs to be exposed to four varieties of packages. A fifth appears optional.
Named after the region in Trinidad it originated, Guaico Culex was discovered during a comprehensive study by the US Army Medical Team to isolate mosquito-borne viruses around the globe. While researchers do not believe Guaico Culex virus can infect mammals, they recently discovered a closely related variety in Uganda’s red colobus monkeys.
#6: Human Endogenous Retrovirus
Roughly 8 percent of the human genome comes from ancient viruses. Retroviruses reproduce by inserting their genetic material into a host and hijacking its replication machinery. Occasionally, these viruses infect sperm and egg cells. If these cells survive, they go on to create an organism containing the virus’s DNA in every cell. These are referred to as endogenous retroviruses—in humans, HERVs. The vast majority are considered non-functional “fossils.” However, a small portion are still intact and can make infectious particles.
Despite being millions of years old, the HERV-K group of viruses appears capable of replicating. Researchers recently discovered a variant that contains no mutations that would downgrade its function. It is believed that this HERV-K remained “alive” within humans until recently. Scientists are unsure whether the dormant virus could reemerge. There is speculation that HERV-K might have been selected for a survival advantage it offered.
#5: Bourbon Virus
A Kansas farmer recently died from a mysterious tick-borne viral infection. The man’s symptoms began with nausea, weakness, and diarrhea. Lung and kidney failure followed. Doctors treated him with antibiotics, the standard course of action for tick-borne illness. Nothing worked. After 10 days in the hospital, he was dead.
With only one confirmed case, doctors are clueless about the disease’s full spectrum. It might be a killer. Or this might be a rare case in which a mild disease became deadly. The best defense is to avoid tick contact by wearing long pants, using insect repellent, and performing frequent tick-checks.
#4: Siberian Giant Virus
A French research team recently unearthed a 30,000-year-old giant virus from the Siberian permafrost—and it’s still infectious. The virus was discovered in a soil sample from 98 feet beneath the ground. It is wider than other giant viruses, and large enough to be seen through a standard microscope.
The team fished for viruses using amoebas, their target hosts, as bait. The amoeba starting dying, and researchers discovered they were laden with these ancient giants. Unlike most viruses, which attack the nucleus, sibericum sets up replication factories in the host’s cytoplasm. Although sibericum only targeted amoeba, another giant virus dubbed Marseillesvirus recent infected an 11-year-old boy in France. It is possible that dangerous viruses also lurk deep underground. More than any other factor, human activities like drilling and mining are likely to unearth these slumbering monsters.
#3: Deep-Sea Virus
Researchers now believe there is more biomass inside earth’s dark, nutrient-deprived depths than anywhere else. In the ocean depths off California, they recently made a remarkable discovery into that mysterious biomass: a virus that infects methane-eating archaea, small bacteria-like organisms, on the ocean floor. Samples were collected from a deep methane seep by pushing tubes into the ocean sediment. Back in the lab, the sediments were fed methane, which triggered archaea growth—along with their viral parasites.
The virus selectively targets one of its own genes for mutation. So do the archaea. The target of the mutations are the tips of the virus, which come into contact with their host. It is a countermeasure against tarchaea’s own selective mutation defenses. This has led to a deep-sea arms race. Partial genetic matches between the California deep-sea viruses and ones discovered around Norway suggest global distribution.
#2: Mysterious Paralysis
In 2015, a wave of American children suffered from acute flaccid paralysis. The outbreak coincided with the flaring up of another respiratory disease caused by the enterovirus EV-D68, a relative of poliovirus. Many suspected a correlation. However, EV-D68 is not known for causing systemic problems like paralysis, and it was only found in 20 percent of the cases. A case from Virginia led some to believe that the cause might be from another virus called C105.
Before the Virginia case, C105 had only been identified in Peru and the Republic of Congo. The disease is associated with respiratory problems. However, a few African cases were linked with paralysis. The C105 theory could explain why 80 percent of the patients tested negative for EV-D68. Yet none of the patients were found to have enterovirus in their spinal fluid, which would account for the neurological symptoms. The cause of the outbreak remains a mystery.
#1: Undiagnosed Hemorrhagic Fever Syndrome
South Sudan is plagued by violence, hunger, and now a mysterious viral outbreak. So far, 10 people have died from Ebola-like symptoms of bleeding, fever, and vomiting. However, Ebola is not the culprit. Doctors have dubbed the disease “undiagnosed hemorrhagic fever syndrome.” Last year, Darfur in Sudan had 129 fatalities from an unidentified illness. It is not yet known whether they are the same disease.
Blood samples for infected patients have revealed a host of viruses: onyong-nyong, chikungunya, and dengue fever. However, none explain the 10 deaths, and none contained Ebola. Most believe this is a virus spread by ticks or mosquitoes, but some are not ruling out the possibility of a bacteria or parasitic origin. So far, there is no evidence of person-to-person transmission, and 75 percent of the victims are under 20. Violent civil war and underdevelopment in the region prevent effective research into the disease’s origins.
————————————————————————–
These 12 Viruses Look Beautiful Up Close But Would Kill You If They Could
January 23, 2014
https://www.huffpost.com/entry/deadly-viruses-beautiful-photos_n_4545309
————————————————————————–
These viruses are the most likely to trigger the next pandemic, according to scientists
April 07, 2021
https://www.livescience.com/spillover-virus-ranking-tool.html
————————————————————————–
New virus found only in Israeli bat with unknown human infection potential
SEPTEMBER 11, 2020
The findings are significant as bats are reservoirs for emerging viruses, and have been attributed to virus outbreaks such as SARS, MERS and many more.
Researchers have found a novel poxvirus on an Egyptian fruit bat in Israel that so far hasn’t been seen anywhere else in the world.
The findings are significant as bats are reservoirs for emerging viruses, and have been attributed to virus outbreaks such as Severe Acute Respiratory Syndrome (SARS), Middle East Respiratory Syndrome (MERS), and many more. Some even attribute the current coronavirus pandemic to bats, although it has not yet been proven.
The poxvirus, called Israel Rousettus aegyptiacus poxvirus (IsrRAPXV), was found on a bat residing in the Zoological Gardens in Tel Aviv. The samples were originally taken from the bat in 2014, however, research has been ongoing for several years; the discovery that the virus only exists in Israel was only made recently in 2018, and the study, ‘A novel poxvirus isolated from an Egyptian fruit bat in Israel,’ was published only this year in Veterinary Medicine and Science.
IsrRAPXV is unique in that not only has it not been detected anywhere else in the world, but it’s also the first time that a poxvirus has been identified in Israel.
The novel poxvirus was discovered after a bat at the Zoological Gardens in Tel Aviv presented with symptoms deemed clinically normal for a poxvirus; consisting of multiple vesicular and nodular skin lesions on the wing membranes. The bat was moved from a captive fruit bat colony, after which scientists took multiple biopsies from the bat before they released it to an open colony with unrestricted access to the external environment.
A multitude of tests were subsequently performed using samples taken from the biopsies, and later, in order to test that what was found was indeed a poxvirus, DNA was extracted. Researchers found that the test results implied that the fruit bat was infected by an unreported and presumably new poxvirus.
Cell culture isolation, histopathology, electron microscopy and molecular analysis revealed the presence of a novel bat poxvirus. More tests were conducted in order to elaborate on previous findings, and in 2018 the Israeli poxvirus was provisionally designated as IsrRAPXV.
The zoonotic potential, otherwise described as the potential for the ability of the virus to jump from bats to humans, is unknown. Bats are reservoirs of numerous viral agents that are transmissible to humans and animals, an example being the Hendra virus (HeV), which can pass from fruit bats to flying foxes, and from horses to humans, according to the government website for New South Wales.
Hendra virus is so named as it was first identified in the Brisbane suburb of Hendra, Australia, 1994. To date total of seven humans have contracted Hendra virus from infected horses, according to the World Health Organization.
Notably similar pox-like lesions that were found on the Egyptian fruit bat were described in an Australian little red flying fox (Pteropus scapulatus) that was trapped in the Kimberley region of North Western Australia. However, DNA was extracted from a lesion sample and the presence of something called PTPV was confirmed.
Other zoonotic viruses that have emerged in humans from bats including the Nipa Virus, which can result in acute respiratory illness and fatal encephalitis, yet only a few known cases have been identified in Asia. In addition, are filoviruses, which can cause severe hemorrhagic fever in humans and nonhuman primates, according to the US Centers for Disease Control.
So far, three genera of this virus family have been identified: Cuevavirus, Marburgvirus and Ebolavirus, the latter two having recently been identified in fruit bats in Africa, the CDC states.
Egyptian fruit bats roost in large colonies that provide many opportunities for disease transmission including deposition of the virus on body surfaces followed by infection of abrasions or auto‐inoculation during grooming, according to the IsrRAPXV study, written by Dan David Irit Davidson Asaf Berkowitz Sharon Karniely Nir Edery Velizar Bumbarov Orly Laskar Ron Elazari‐Volcani.
It adds that although “no pox infections have been reported in humans exposed to this bat, it would be prudent for professionals working with bats to wear protective clothes and gloves when handling them.”
https://www.jpost.com/health-science/new-virus-found-only-in-israeli-bat-with-unknown-human-infection-potential-641932
————————————————————————–
Novel Coronavirus Discovered in British Bats – Related to the Virus That Causes COVID-19
July 20, 2021
https://scitechdaily.com/novel-coronavirus-discovered-in-british-bats-related-to-the-virus-that-causes-covid-19/
————————————————————————–
Identification of novel bat coronaviruses sheds light on the evolutionary origins of SARS-CoV-2 and related viruses
2021
https://pubmed.ncbi.nlm.nih.gov/34147139/
————————————————————————–
Cospeciation of coronavirus and paramyxovirus with their bat hosts in the same geographical areas
2021
https://pubmed.ncbi.nlm.nih.gov/34325659/
————————————————————————–
Detection of potentially novel paramyxovirus and coronavirus viral RNA in bats and rats in the Mekong Delta region of southern Viet Nam
2017
https://pubmed.ncbi.nlm.nih.gov/28418192/
————————————————————————–
Coronavirus and paramyxovirus in bats from Northwest Italy
2017
https://pubmed.ncbi.nlm.nih.gov/29273042/
————————————————————————–
Paramyxo- and Coronaviruses in Rwandan Bats
2019
https://www.mdpi.com/2414-6366/4/3/99/pdf
————————————————————————–
A Recently Discovered Pathogenic Paramyxovirus, Sosuga Virus, is Present in Rousettus aegyptiacus Fruit Bats at Multiple Locations in Uganda.
28 Apr 2015
http://europepmc.org/article/PMC/5022529
————————————————————————–
Achimota Pararubulavirus 3: A New Bat-Derived Paramyxovirus of the Genus Pararubulavirus.
30 Oct 2020
https://europepmc.org/article/MED/33143230
————————————————————————–
Zoonotic Vectorborne Pathogens and Ectoparasites of Dogs and Cats in Eastern and Southeast Asia
2020
https://wwwnc.cdc.gov/eid/article/26/6/19-1832_article
————————————————————————–
Arthropod Ectoparasites Have Potential to Bind SARS-CoV-2 via ACE
2021
Coronavirus-like organisms have been previously identified in Arthropod ectoparasites (such as ticks and unfed cat flea). Yet, the question regarding the possible role of these arthropods as SARS-CoV-2 passive/biological transmission vectors is still poorly explored. In this study, we performed in silico structural and binding energy calculations to assess the risks associated with possible ectoparasite transmission. We found sufficient similarity between ectoparasite ACE and human ACE2 protein sequences to build good quality 3D-models of the SARS-CoV-2 Spike:ACE complex to assess the impacts of ectoparasite mutations on complex stability. For several species (e.g., water flea, deer tick, body louse), our analyses showed no significant destabilisation of the SARS-CoV-2 Spike:ACE complex, suggesting these species would bind the viral Spike protein. Our structural analyses also provide structural rationale for interactions between the viral Spike and the ectoparasite ACE proteins. Although we do not have experimental evidence of infection in these ectoparasites, the predicted stability of the complex suggests this is possible, raising concerns of a possible role in passive transmission of the virus to their human hosts.
Human respiratory viruses continue to spread in wild chimpanzees
January 19, 2019
https://ghi.wisc.edu/human-respiratory-viruses-continue-to-spread-in-wild-chimpanzees/
————————————————————————–
Chimps are now dying of the common cold and they are all at risk
25 January 2018
https://www.newscientist.com/article/2159509-chimps-are-now-dying-of-the-common-cold-and-they-are-all-at-risk/
————————————————————————–
Fatal septic shock due to Capnocytophaga canimorsus bacteremia masquerading as COVID-19 pneumonia – a case report
2021
Abstract
Capnocytophaga canimorsus (C. canimorsus) infections are rare and usually present with unspecific symptoms, which can eventually end in fatal septic shock and multiorgan failure. The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) related coronavirus disease 2019 (COVID-19), on the other hand, is predominantly characterized by acute respiratory failure, although other organ complications can occur. Both infectious diseases have in common that hyperinflammation with a cytokine storm can occur. While microbial detection of C. canimorsus in blood cultures can take over 48 h, diagnosis of SARS-CoV-2 is facilitated by a widely available rapid antigen diagnostic test (Ag-RDT) the results of which are available within half an hour. These Ag-RDT results are commonly verified by a nucleic acid amplification test (NAAT), whose results are only available after a further 24 h.
https://pubmed.ncbi.nlm.nih.gov/34344315/
————————————————————————–
Woman Who Thought Who Thought She Had COVID Actually Caught Typhus from A Dead Rat
8-4-2021
https://www.newsweek.com/woman-thought-had-covid-actually-caught-typhus-dead-rat-symptoms-1616169
————————————————————————–
Diseases That Can Spread Between Animals and People
https://www.cdc.gov/healthypets/diseases/index.html
————————————————————————–
A COVID-19 vaccine strategy to give the body ‘border protection’
Aug 11, 2021
Study in animals shows a way to promote immune response in nose, mouth and blood
https://news.osu.edu/a-covid-19-vaccine-strategy-to-give-the-body-border-protection/
————————————————————————–
Direct detection of SARS-CoV-2 antisense and sense genomic RNA in human saliva by semi-autonomous fluorescence in situ hybridization: A proxy for contagiousness?
August 17, 2021
Abstract
Saliva is a matrix which may act as a vector for pathogen transmission and may serve as a possible proxy for SARS-CoV-2 contagiousness. Therefore, the possibility of detection of intracellular SARS-CoV-2 in saliva by means of fluorescence in situ hybridization is tested, utilizing probes targeting the antisense or sense genomic RNA of SARS-CoV-2. This method was applied in a pilot study with saliva samples collected from healthy persons and those presenting with mild or moderate COVID-19 symptoms. In all participants, saliva appeared a suitable matrix for the detection of SARS-CoV-2. Among the healthy, mild COVID-19-symptomatic and moderate COVID-19-symptomatic persons, 0%, 90% and 100% tested positive for SARS-CoV-2, respectively. Moreover, the procedure allows for simultaneous measurement of viral load (‘presence’, sense genomic SARS-CoV-2 RNA) and viral replication (‘activity’, antisense genomic SARS-CoV-2 RNA) and may yield qualitative results. In addition, the visualization of DNA in the cells in saliva provides an additional cytological context to the validity and interpretability of the test results. The method described in this pilot study may be a valuable diagnostic tool for detection of SARS-CoV-2, distinguishing between ‘presence’ (viral load) and ‘activity’ (viral replication) of the virus. Moreover, the method potentially gives more information about possible contagiousness.
https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0256378
————————————————————————–
Vaccine made of mosquito spit could help stop next epidemic
June 12, 2020
https://nypost.com/2020/06/12/how-a-vaccine-made-of-mosquito-spit-could-help-stop-the-next-epidemic/
————————————————————————–
How a vaccine made of mosquito spit could help stop the next epidemic
June 11, 2020
https://www.yahoo.com/news/vaccine-made-mosquito-spit-could-223511907.html
————————————————————————–
Mosquito saliva vaccine? NIH tests new way to fight illness
February 21, 2017
https://www.foxnews.com/health/mosquito-saliva-vaccine-nih-tests-new-way-to-fight-illness
————————————————————————–
The Future of Mini Drones or MAV’s (Micro Air Vehicles)
2017

https://blog.prv-engineering.co.uk/future-micro-drones/
————————————————————————–
Does This Photo Show a Robotic Spy Mosquito?
September 10, 2015
http://wafflesatnoon.com/does-this-photo-show-a-robotic-spy-mosquito/
————————————————————————–
Could a Single Vaccine Prevent Multiple Diseases Spread by Mosquitoes?
June 12, 2018
https://www.acsh.org/news/2018/06/12/could-single-vaccine-prevent-multiple-diseases-spread-mosquitoes-13073
————————————————————————–
Kanyawara Virus: A Novel Rhabdovirus Infecting Newly Discovered Nycteribiid Bat Flies Infesting Previously Unknown Pteropodid Bats in Uganda
2017
https://pubmed.ncbi.nlm.nih.gov/28706276/
————————————————————————–
Conference Paper: Identification Of Host Dependency Factors For Mers-cov Infection By Genome-wide Crispr-cas9 Screen
2019
http://hub.hku.hk/handle/10722/274155
Abstract
Human infection of Middle East Respiratory Syndrome Coronavirus (MERS-CoV) was first reported in 2012 and two large-scale outbreaks took place in Saudi Arabia (2014) and South Korea (2015). From 2012 through the end of 2018, 2279 cases were reported to WHO, with a fatality rate of 35% (806 deaths). However, no specific antiviral or vaccine against MERS-CoV infection is currently available. Further investigation of virus‑host interaction may provide mechanistic insights into the development of therapeutic agents. In this study, we performed a genome-wide CRISPR-Cas9 screen to identify host dependency factors for MERS-CoV infection in a cultured cell model. From this positive-selection screen, we identified 13 host protein factors while one of them is the known receptor (DPP4) for the MERS-CoV. Next, we characterized one of these host dependency factors, PTBP1, and investigated its role in the MERS-CoV infection. Interestingly, we found that PTBP1 does not directly regulate the MERS-CoV viral replication, but limits the viral entry through the negative regulation of DPP4 receptor expression. Therefore targeting PTBP1 expression may help to restrict the MERS-CoV infection through the downregulation of its receptor DPP4. This study was supported by Research Grant Council–Theme‑based Research Scheme (T11-707/15-R).
——————————
Reverse genetics with a full-length infectious cDNA of the Middle East respiratory syndrome coronavirus
2013
https://www.pnas.org/content/110/40/16157
—————————
Assembly of a MERS-CoV full-length cDNA clone as a BAC.
https://www.researchgate.net/figure/Assembly-of-a-MERS-CoV-full-length-cDNA-clone-as-a-BAC-A-Genome-organization-of-the_fig1_256491232
—————————
Coronaviruses and arteriviruses display striking differences in their cyclophilin A-dependence during replication in cell culture
April 2018
https://www.sciencedirect.com/science/article/pii/S0042682217304014
————————–
News Feature: Avoiding pitfalls in the pursuit of a COVID-19 vaccine
March 30, 2020
https://www.pnas.org/content/early/2020/03/27/2005456117
As they race to devise a vaccine, researchers are trying to ensure that their candidates don’t spur a counterproductive, even dangerous, immune system reaction known as immune enhancement.
The teams of researchers scrambling to develop a coronavirus disease 2019 (COVID-19) vaccine clearly face some big challenges, both scientific and logistical. One of the most pressing: understanding how the immune system interacts not only with the pathogen but with the vaccine itself—crucial insights when attempting to develop a safe and effective vaccine.
——————————-
Synthetic Biologists Think They Can Develop a Better Coronavirus Vaccine Than Nature Could
March 9, 2020
https://www.scientificamerican.com/article/synthetic-biologists-think-they-can-develop-a-better-coronavirus-vaccine-than-nature-could/
The Bill and Melinda Gates Foundation and the National Institutes of Health are betting on a different approach than existing efforts for battling COVID-19
Even as companies rush to develop and test vaccines against the new coronavirus, the Bill and Melinda Gates Foundation and the National Institutes of Health are betting that scientists can do even better than what’s now in the pipeline.
If, as seems quite possible, the Covid-19 virus becomes a permanent part of the world’s microbial menagerie rather than being eradicated like the earlier SARS coronavirus, next-gen approaches will be needed to address shortcomings of even the most cutting-edge vaccines: They take years to develop and manufacture, they become obsolete if the virus evolves, and the immune response they produce is often weak.
With Gates and NIH funding, the emerging field of synthetic biology is answering the SOS over Covid-19, aiming to engineer vaccines that overcome these obstacles. “It’s all of us against the bug,” said Neil King of the University of Washington, who has been part of the hunt for a coronavirus vaccine since 2017.
Although the Gates Foundation is spreading its bets among several cutting-edge vaccine platforms, including those using genetic material, one based on synthetic biology has real promise. “We may need an approach that can get you millions and even billions of doses,” said immunologist and physician Lynda Stuart, who directs the foundation’s vaccine research. Gates announced last month that it will funnel $60 million to Covid-19 research, including vaccines.
A vaccine created through the tinkering of synbio looks not only scalable to a level of billions but also like it will work without the need for refrigeration. All that, Stuart said, “will be super important to protect people from coronavirus who are otherwise left behind, such as those in sub-Saharan Africa.”
King and his synbio colleagues knew there would be another coronavirus epidemic, like the SARS and MERS outbreaks before this one, he said, “and there will be another one after this,” perhaps from yet another member of this virus family. “We need a universal coronavirus vaccine.”
Achieving that is so high on scientists’ to-do list that when President Trump visited NIH last week, his tour included the lab that’s collaborating with UW’s, and researchers showed him a mock-up of what synthetic biology can do: Design and build nanoparticles out of proteins and attach viral molecules in a repetitive array so that, when the whole thing is packed into a vaccine, it can make people resistant to the new coronavirus. (The human immune system has evolved to interpret repetitive arrangements of molecules as a sign of danger: bacterial cell walls have repetitive chemical groups on them.)
With a few tweaks, the nanoparticle can be studded with molecules from additional coronaviruses to, scientists hope, protect against all of them—the original SARS virus, MERS, and, crucially, a mutated form of the Covid-19-causing virus, called SARS-CoV-2.
Even compared to the DNA and RNA vaccines against Covid-19 that Moderna Therapeutics, CureVac, and Inovio Pharmaceuticals are speeding toward human testing, the synbio approach has advantages. These companies’ experimental vaccines contain synthetic (that is, lab-made) strands of RNA or DNA that code for protein molecules on the virus’s surface. Once the vaccine delivers the genetic material into cells, the cells follow the genetic instructions to churn out the viral protein. The idea is that the body would see that as foreign, generate antibodies to it, and if all goes well thereby acquire immunity to the virus. But safety tests of mRNA vaccines have turned up adverse events, and it’s not clear how potent they’ll be. Moderna plans to begin safety testing in healthy volunteers next month.
With all due respect to nature, synthetic biologists believe they can do better. Using computers, they are designing new, self-assembling protein nanoparticles studded with viral proteins, called antigens: these porcupine-like particles would be the guts of a vaccine. If tests in lab animals of the first such nanoparticle vaccine are any indication, it should be more potent than either old-fashioned viral vaccines like those for influenza or the viral antigens on their own (without the nanoparticle).
The first step toward the molecule that was presented to Trump is to “play Legos with proteins,” as King put it.
That starts with the nanoparticle—the body of the porcupine. Its shape and composition must be such that the protein’s building blocks not only spontaneously self-assemble and stick together but also turn into something that can display the viral antigens in a way the immune system will strongly respond to. Using a computational protein-design algorithm, scientists might determine that, for instance, a nanoparticle 25 nanometers across and made of 60 identical pieces is ideal for presenting the antigens sotheir most immunity-inducing side faces outward, where the immune system can most easily “see” it.
“We might try 1 million variants on the computer” before finding the optimal shape and protein composition, meaning which protein sequence will spontaneously form the ideal nanoparticle, King said.
The next step is to take lab-made DNA that codes for the designed protein, stick it into E. coli bacteria, and wait for the bugs to follow the genetic instructions, manufacturing the desired protein like a tiny, living assembly line. Extracted from the bacteria, purified, and mixed together in a test tube, the proteins spontaneously self-assemble into the bespoke nanoparticle.
“When it works, we get exactly the protein we designed by computer, with every atom where we want it,” King said.
The next step is to stick the quills onto the porcupine. For the virus that causes Covid-19, the quills are the “spike protein,” a molecule that fits into receptors on cells and ushers the virus inside. Scientists led by UW’s David Baker predicted the structure of this antigen from the virus’s genome, and scientists at the University of Texas, Austin, and NIH confirmed it with a Nobel-winning form of electron microscopy.
King and his colleagues then scrutinize the spike protein to see which part of it might work best in a vaccine and how to position multiple copies of it. “It turns out that if you stick 20 of them onto your nanoparticle in an ordered, repetitive array, you can get a stronger immune response than with the [spike] protein alone,” Baker said—another reason why the nanoparticle approach might prove more effective than RNA and DNA vaccines. NIH and the UW groups have begun testing the antigen-studded nanoparticles in mice to see what kind of immune response they trigger.
Making nanoparticles the core of a vaccine “does a number of useful things,” Stuart said. It reduces or eliminates the need for an adjuvant, an ingredient that boosts the immune response; the nanoparticle is enough on its own. Sticking antigens on it makes the whole complex so tolerant of heat (“you could almost boil it,” Stuart said) that refrigeration isn’t necessary, a crucial feature for vaccines to be deployed in resource-poor countries. And because the nanoparticle can be studded with antigens from several viruses, she said, “you could get a pan-coronavirus vaccine.”
They’re cautiously optimistic because of a recent success. An experimental vaccine against respiratory syncytial virus (RSV), the main cause of pneumonia in children, is also made of a computer-designed nanoparticle that self-assembles from protein building blocks and is studded with an engineered version of RSV’s key antigen. When tested in mice and monkeys, it produced 10 times more antibodies than an experimental RSV vaccine based on traditional technology, King’s team reported last year in Cell. The Seattle biotech start-up Icosavax is moving the vaccine toward clinical trials. (King chairs its scientific advisory board.)
It was the first time the structure and other characteristics of an antigen had been designed at the atomic level and incorporated into a vaccine, scientists at vaccine giant GSK wrote, hailing the work as “a quantum leap” in vaccine design.
The Gates Foundation, in addition to supporting the research, is working to pair the scientists with manufacturers, Stuart said: “We want to identify the people who can manufacture these at scale.”
—————————————————
Future Defenses Against Colorectal Cancer May Include a Vaccine and Genetic Engineering
March 2, 2020
https://www.cancer.org/latest-news/future-defenses-against-colorectal-cancer-may-include-a-vaccine-and-genetic-engineering.html
—————————————————-
—————————————————-
A Case for the Ancient Origin of Coronaviruses
2013
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3676139/
Abstract
Coronaviruses are found in a diverse array of bat and bird species, which are believed to act as natural hosts. Molecular clock dating analyses of coronaviruses suggest that the most recent common ancestor of these viruses existed around 10,000 years ago. This relatively young age is in sharp contrast to the ancient evolutionary history of their putative natural hosts, which began diversifying tens of millions of years ago. Here, we attempted to resolve this discrepancy by applying more realistic evolutionary models that have previously revealed the ancient evolutionary history of other RNA viruses. By explicitly modeling variation in the strength of natural selection over time and thereby improving the modeling of substitution saturation, we found that the time to the most recent ancestor common for all coronaviruses is likely far greater (millions of years) than the previously inferred range.
INTRODUCTION
Coronaviruses (family Coronaviridae, subfamily Coronavirinae) are important pathogens of birds and mammals. Coronaviruses are positive-sense RNA viruses and are currently classified into four genera: Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus. Alphacoronaviruses and betacoronaviruses are found exclusively in mammals, whereas gammacoronaviruses and deltacoronaviruses primarily infect birds. The identification of severe acute respiratory syndrome (SARS) coronavirus in 2003 prompted an intensive search for novel coronaviruses, resulting in the detection of a number of novel coronaviruses in humans, domesticated animals, and wildlife (3–7). Interestingly, surveillance of coronaviruses in wild animals has led to the discovery of the greatest diversity of coronaviruses in bat and avian species, which suggests that these animals are the natural reservoirs of the viruses. Indeed, phylogenetic studies of bat and avian coronaviruses suggest an ancient relationship with possible codivergence and coevolution with their hosts. Conversely, many coronaviruses found in bats and other mammals diverged near the tips of coronavirus phylogeny, suggesting that these viruses were the result of recent cross-species transmission events.
Molecular clock analysis based on the RNA-dependent RNA polymerase (RdRp) genomic region suggests a time of most recent common ancestor (tMRCA) for the four coronavirus genera of around 10,100 years ago, with a mean rate of 1.3 × 10−4 nucleotide substitutions per site per year. This tMRCA estimate is difficult to reconcile with a hypothetical ancient, coevolutionary relationship between coronaviruses and their bat or bird hosts. Moreover, a group of genetically related, yet distinct, alphacoronaviruses have been detected in different mouse-eared bats (Myotis spp.) on multiple continents. However, these bat species do not migrate long distances, with few traveling farther than hundreds of miles to overwinter sites (Gary F. McCracken, personal communication). Yet, the tMRCA of Alphacoronavirus is estimated to be around 200 or 4,400 years ago, on the basis of analyses of helicase (9) and RdRp (10), respectively. The limited interaction among these bat populations suggests a more ancient evolutionary association with alphacoronaviruses (i.e., codivergence or coevolution), which is incompatible with the relatively young viral tMRCAs. Notably, coronaviruses have a unique proofreading mechanism for viral RNA replication (3, 14); because of the exoribonuclease activity of viral nonstructural protein 14 (Nsp14), the mutation rate of coronaviruses has been found to be similar to that of single-stranded DNA viruses (∼1 × 10−5 to 1 × 10−6 mutation per site per replication) and well below those measured in other RNA viruses (∼1 × 10−3 to 1 × 10−5 mutation per site per replication). For these reasons, we speculate that there is a substantial underestimation of the length of the natural evolutionary history of coronaviruses.
Recent developments in the modeling of the evolution of RNA viruses have revealed that purifying selection can mask the ancient age of viruses that appear to have recent origins, according to a molecular clock (e.g., measles, Ebola, and avian influenza viruses). Strong purifying selection can maintain evidence of sequence homology long after saturation has occurred at synonymous sites; this phenomenon can lead to underestimation of the overall depth of a viral phylogeny. In the absence of strong purifying selection, nucleotide sequences would diverge more quickly, lose detectable homology, and become difficult to align and compare. Here, we asked if similar evolutionary patterns led to underestimation of the tMRCA of the coronavirus lineage. We employed evolutionary models that account for variation in the pressure of natural selection across sites in viral loci and lineages in their phylogenies. Our results indicate that coronaviruses are orders of magnitude older than suggested by previous molecular clock analyses.
MATERIALS AND METHODS
Sequence data sets.
Representative coronavirus genomes (n = 43; Table 1) from the four genera were selected for this study. For several viral taxa, genomic sequences from multiple sampling years were included, though these dates were not used explicitly in our analyses. To avoid highly recombinant sequences and/or sequence misalignments, we specifically selected partial viral sequences from five relatively conserved genomic regions (Fig. 1 and Table 1): Nsp15-16 (1,320 nucleotides [nt]), the matrix protein (640 nt), papain-like protease 2 (PLP2; 620 nt), the RdRp (1,860 nt), and the Y domain (400 nt) (18). These nucleotide sequences were aligned at the amino acid level by MUSCLE (19) as described previously.
Table 1
Coronavirus sequences analyzed in this study
Genus and virus name Host species Strain Accession no. Sampling yr
Alphacoronavirus
Bat coronavirus 1Aa Miniopterus magnater AFCD62 EU420138 2005
Bat coronavirus 1Ba Miniopterus pusillus AFCD307 EU420137 2006
Bat coronavirus HKU2 Rhinolophus sinicus HKU2/GD/430/2006 EF203064 2006
Bat coronavirus HKU8 Miniopterus pusillus AFCD77 EU420139 2005
Feline coronavirusb Felis catus UU2 FJ938060 1993
Feline coronavirusb Felis catus UU54 JN183883 2010
Human coronavirus NL63 Homo sapiens NL63/DEN/2009/14 JQ765564 2009
Human coronavirus NL63 Homo sapiens NL63/DEN/2005/1876 JQ765575 2005
Porcine epidemic diarrhea virus Sus scrofa CH/FJND-3 JQ282909 2011
Porcine epidemic diarrhea virus Sus scrofa CH/S JN547228 1986
Transmissible gastroenteritis virusb Sus scrofa H16 FJ755618 1973
Transmissible gastroenteritis virusb Sus scrofa Purdue DQ811789 1952
Betacoronavirus
Bat coronavirus HKU5 Pipistrellus abramus TT07f EF065512 2006
Bat coronavirus HKU9 Rousettus leschenaulti 10-1 HM211100 2006
Bat coronavirus/133/2005 Tylonycteris pachypus BtCoV/133/2005 DQ648794 2005
Bat SARS coronavirusc Rhinolophus pearsoni Rp3 DQ071615 2004
Bovine coronavirusd Bos taurus DB2 DQ811784 1983
Bovine coronavirusd Bos taurus E-AH187-TC FJ938064 2000
Civet SARS coronavirusc Paguma larvata SZ3 AY304486 2003
Human coronavirus HKU1 Homo sapiens Caen1 HM034837 2005
Human coronavirus OC43d Homo sapiens HK04-02 JN129835 2004
Human SARS coronavirusc Homo sapiens HKU-39849 isolate UOB JQ316196 2003
Mouse hepatitis virus Mus musculus MHV-MI AB551247 1994
Mouse hepatitis virus RA59/R13 Mus musculus RA59/R13 FJ647218 2006
Sammbar deer coronavirus Cervus unicolor US/OH-WD388-TC FJ425188 1994
Gammacoronavirus
Duck coronavirus Duckf DK/CH/HN/ZZ2004 JF705860 2004
Infectious bronchitis viruse Gallus gallus Holte GU393336 1954
Infectious bronchitis viruse Gallus gallus ck/CH/LHLJ/100902 JF828980 2010
Infectious bronchitis viruse Gallus gallus Conn46 FJ904719 1991
Infectious bronchitis viruse Gallus gallus Mass41 FJ904721 1972
Infectious bronchitis viruse Gallus gallus Massachusetts GQ504724 1941
Turkey coronaviruse Meleagris gallopavo IN-517 GQ427175 1994
Turkey coronaviruse Meleagris gallopavo TX-1038 GQ427176 1998
Turkey coronaviruse Meleagris gallopavo VA-74 GQ427173 2003
Deltacoronavirus
Bulbul coronavirus HKU11 Pycnonotus jocosus HKU11-934 FJ376619 2007
Common-moorhen coronavirus HKU21 Gallinula chloropus HKU21-8295 JQ065049 2007
Magpie robin coronavirus HKU18 Copsychus saularis HKU18-chu3 JQ065046 2007
Munia coronavirus HKU13 Lonshura striata HKU13-3514 FJ376622 2007
Night-heron coronavirus HKU19 Nycticorax nycticorax HKU19-6918 JQ065047 2007
Sparrow coronavirus HKU17 Passer montanus HKU17-6124 JQ065045 2007
Thrush coronavirus HKU12 Turdus hortulorum HKU12-600 FJ376621 2007
White-eye coronavirus HKU16 Zosterops sp. HKU16-6847 JQ065044 2007
Wigeon coronavirus HKU20 Anas penelope HKU20-9243 JQ065048 2008
aViruses are classified as a single species (Miniopterus bat coronavirus 1).
bViruses are classified as a single species (Alphacoronavirus 1).
cViruses are classified as a single species (Severe acute respiratory syndrome-related coronavirus).
dViruses are classified as a single species (Betacoronavirus 1).
eViruses are classified as a single species (Avian coronavirus).
fHost species cannot be determined.
Phylogenetic analyses.
Each of the five target sequences was screened for recombination with a genetic algorithm tool, GARD (21). Maximum-likelihood phylogenies were constructed for each nonrecombinant region with PHYML 3.0 (22), implemented in Seaview 4.0 (23), with a subtree-pruning and regrafting search algorithm and the general time-reversible substitution model with a four-bin gamma rate distribution (GTR + Γ4).
With these topologies, branch lengths were re-estimated in HyPhy (24) under GTR + Γ4 and a branch site random effects likelihood (BS-REL) model (25). The BS-REL model was implemented to account for the effects of variable selection pressure across codon sites and phylogenetic lineages by inferring three unconstrained ω classes (dN/dS: nonsynonymous substitution rate/synonymous substitution rate) for each branch and estimating the proportion of sites in the alignment that evolved under each ω class.
To estimate the variance in branch length estimates produced by the BS-REL model, we used a modified Latin hypercube sampling (LHS) importance resampling scheme (M = 500 samples), described in detail previously. LHS is a standard technique that can be efficiently parallelized, an important consideration here. It is used to assess the variability of high-dimensional distributions by discretizing the volume of parameter space around the maximum-likelihood estimate into N bins (10,000 in our case) of approximately equal probability along each coordinate and sampling the bin for each parameter in a way that no two parameters share the same bin index (this technique maximizes space coverage). Samples are reweighted according to their likelihood, and resamples (M = 500) are drawn from this distribution. A modified version of LHS, which we use here, has also been shown to compare favorably to other techniques for the assessment of parameter uncertainty.
RESULTS
Recombination detection.
Phylogenies with interpretable branch lengths can be inferred only when analyzing nonrecombinant regions. Therefore, we screened each of the five highly conserved coronavirus target regions with GARD (21). Within the Nsp15-16 region, two recombination breakpoints were detected, and phylogenetic incongruity was confirmed by a Kishino-Hasegawa test (P < 0.01) (27, 28). The first nonrecombinant section encompassed Nsp15 and 80 nt of Nsp16, which we refer to as Nsp15+; the second section fell entirely within Nsp16. Because of its short length (300 nt), the third section was not included in later analyses. Within the RdRp region, a single recombination breakpoint was detected with GARD, but it is unlikely that this breakpoint represents a true recombination event since the two topologies were not significantly different according to the Kishino-Hasegawa test. This putative breakpoint detected in the RdRp region was likely due to elevated rate variation across the sequence. No recombination breakpoints were detected in the other three loci.
The maximum-likelihood phylogenies inferred from the six nonrecombinant loci were almost all significantly different from each other (Shimodaira-Hasegawa test, P < 0.05) (29). This finding suggests that the different regions of the coronavirus genome have distinct evolutionary histories and should not be treated as a single region in a phylogenetic analysis. The lone exception was the comparison of the topologies from RdRp and Nsp15+ (P = 0.131). Nevertheless, we chose be conservative and analyze each of the six loci independently.
Branch length expansion.
We employed two models of nucleotide sequence evolution (GTR + Γ4 and BS-REL) to re-estimate the branch lengths of the inferred maximum-likelihood phylogenies. GTR + Γ4 is a standard nucleotide substitution model that has been commonly in many evolutionary virology studies. In contrast, the BS-REL model is a codon model that explicitly accounts for variation in selection pressures across sites and lineages. Both evolutionary models produced comparable length estimates for shorter branches (≤0.05 substitution per site in Fig. 2). However, compared to results inferred from BS-REL, long branches in the coronavirus phylogenies were underestimated by GTR + Γ4 (Fig. 2). Notably, there were a substantial number of long branches in which the number of substitutions per site approached saturation in the BS-REL model, ranging from 8 branches in Nsp16 to 16 branches in the Y domain (Table 2). The lengths of these saturated branches cannot be reliably estimated, although a meaningful lower bound on their lengths can be obtained (e.g., with LHS; see below).

Fig 2
Branch length expansion under the BS-REL model compared to that under the GTR + Γ4 model for six coronavirus sequences: Nsp15+, Nsp16, matrix protein, PLP2, RdRp, and the Y domain. Each point represents a branch in the coronavirus phylogeny. For ease of visualization, BS-REL branch lengths experiencing extreme saturation (>50 substitutions per site) are depicted with infinite length.
Table 2
Tree lengths generated under GTR+Γ4 and BS-REL evolutionary models
Viral sequence Mean dN/dSa No. of branches approaching saturation under BS-RELb Proportion of sites under pervasive purifying selectionc Tree length (no. of substitutions/site) Expansion under BS-REL 95% CI expansion under BS-RELd (lower-upper)
GTR+Γ4 BS-REL
Nsp15+ 0.17 12 0.77 11.3 10,530 935 659–100,725
Nsp16 0.12 8 0.85 10.6 6,488 610 439–157,128
Matrix 0.19 9 0.67 14.8 38,287 2,581 1,868–162,126
PLP2 0.26 13 0.70 15.3 67,831 4,432 3,261–166,920
RdRp 0.11 14 0.87 8.0 230,399 28,860 18,699–48,256
Y domain 0.21 16 0.85 13.3 362,318 27,253 16,504–27,434
aInferred by a single-likelihood ancestral counting method (50).
bOut of a total of 83 (40 internal) branches in the coronavirus phylogeny.
cInferred by a fixed-effects likelihood method (50).
dCIs were estimated by an LHS importance resampling scheme (M = 500 samples).
Among the six nonrecombinant regions, there was substantial variation in the total tree length expansion between GTR + Γ4 and BS-REL, ranging over 3 orders of magnitude (Table 2). Differing selective pressures along lineages among loci appear to account for this variation, as we observed a correlation between the strength of purifying selection along a branch (i.e., lower mean ω) and the relative increase in branch length under BS-REL compared to GTR + Γ4 (Spearman’s nonparametric correlation, P ≤ 0.0001; branches in which either model inferred a length of zero substitutions per site were excluded from this analysis). Thus, lineages that bear the mark of stronger purifying selection generally experienced a greater increase in length under the BS-REL model. Moreover, the number of branches approaching saturation was correlated with a greater expansion in the total length under BS-REL (Spearman’s nonparametric correlation, P < 0.05). Interestingly, simpler, site-based measurements of selection did not correlate with total tree length expansion (e.g., mean dN/dS [P = 0.87] and number of sites of pervasive purifying selection [P = 0.46]), though analyzing only six loci limits the statistical power of these tests. Nonetheless, the relationship between ω and branch length expansion supports the hypothesis that increased purifying selection is responsible for the underestimation of the lengths of long branches in RNA virus phylogenies.
The variance in branch length expansion from GTR + Γ4 to BS-REL, as approximated with LHS, differed among coronavirus loci (Table 2). For the Nsp15+, Nsp16, matrix protein, and PLP2 loci, the upper and lower 95% confidence limits differed by orders of magnitude, suggesting highly imprecise estimates of branch length expansion in these loci. This lack of precision is not surprising, given the extent to which these loci have experienced saturation along their phylogeny. Once a site becomes saturated with repeated substitutions, it is very difficult for an evolutionary model to determine the exact number of substitutions that have taken place. Therefore, the maximum-likelihood estimate will be imprecise. Nevertheless, these results clearly suggested that there is substantial underestimation of the evolutionary history of coronaviruses.
To examine the effects of saturation and determine the limit of reliable divergence inference with BS-REL, we simulated sequence alignments in HyPhy (24) under the maximum-likelihood parameter values from the BS-REL model fitted to the RdRp locus. HyPhy scripts and input files needed to perform the simulations can be downloaded at https://github.com/veg/pubs/tree/master/SARS. The tree inferred by using this locus experienced a dramatic expansion in a previous BS-REL analysis. For example, a single internal branch at this locus accounted for >70% of the total inferred tree length, but it also experienced incredibly strong purifying selection (dN/dS = 0) at 97.5% of the sites and very strong diversifying selection (dN/dS > 100) at the remaining 2.5% of the sites. In the simulations (n = 100), we kept the relative lengths of branches fixed and scaled the entire tree length from 1 to 10,000 expected substitutions/site. We then fitted the GTR + Γ4 and BS-REL models to the replicates to evaluate the saturation behavior of the models. There were striking differences between the models (Fig. 3). GTR + Γ4 consistently, and progressively more badly, underestimated the simulated tree length, starting at the lowest simulated values; this behavior is due to a long internal branch predominantly subject to strong purifying selection, which poses a challenge for the nucleotide-based model (30). Note that this type of saturation does not lead to infinite branch lengths because a majority of the sites are maintained by strong purifying selection. In contrast, the BS-REL model did not saturate until the total length of the tree was on the order of 1,000 substitutions per site. Interestingly, BS-REL estimates are biased slightly upward, largely because of the saturation of nonsynonymous substitutions at positively selected sites and corresponding infinite values of dN/dS.
tMRCA extrapolation.
The current best estimate of the tMRCA of all four coronavirus genera of 10,141 years ago was obtained by Woo et al. (10) by using the RdRp locus; their tMRCA estimate is the only one that includes all four coronavirus genera. Notably, their tMRCA estimates for alpha- and betacoronaviruses are similar to the dates inferred by a similar methodology from a different data set (31). Our results suggest that, despite the comprehensive nature of these studies, there are intrinsic shortcomings in the standard evolutionary models used to estimate branch lengths and the tMRCA of ancient viral lineages like coronaviruses. Nevertheless, Woo et al. calibrated the coronavirus molecular clock with serially sampled sequences, which rely predominantly on the relationship between the branches near the tips of the phylogeny to estimate substitution rates. Notably, it is possible to infer consistent RNA virus substitution rates from these types of serially sampled sequences, even if the tMRCA of older lineages deep in the tree cannot be reliably estimated because of saturation along internal branches (e.g., simian immunodeficiency virus/human immunodeficiency virus [32, 33] and avian influenza virus).
When variation in selection pressure was taken into account using BS-REL, we observed a dramatic expansion of the total length of the RdRp phylogeny: 28,860.4-fold (95% confidence interval [CI], 18,695.7-fold to 48,256.0-fold) (Table 2). If one were to extrapolate the evolutionary ages estimated by Woo et al. to the BS-REL phylogeny, the adjusted tMRCA of coronaviruses would be 293 (95% CI, 190 to 489) million years ago, 4 orders of magnitude greater than that previously inferred. The robustness of this tMRCA estimate is highly dependent on both the rate of evolution estimated by Woo et al. (10), which is inferred primarily from short branches near the tips of the phylogeny, and the accuracy of the new branch lengths, which appear extremely variable. Nevertheless, even with the conservative lower bounds of the tMRCA of coronaviruses and branch length expansion (974 BCE [10] and 18,695.7-fold expansion [Table 2]), the adjusted coronavirus tMRCA of 55.8 million years ago would still be 3 orders of magnitude greater than the current estimate.
DISCUSSION
Our results indicate that the evolutionary history of coronaviruses likely extends much further back in time than previous estimates have suggested. Across the coronavirus genome, there is evidence that standard nucleotide models underestimate the amount of evolution that has occurred by orders of magnitude; strong purifying selection had masked the evidence of thousands or millions of years of evolution in the coronavirus phylogeny. Like many other DNA and RNA viruses—including herpesviruses, lentiviruses, bornaviruses, filoviruses, and foamy viruses —coronaviruses appear to be an ancient viral lineage. Furthermore, our results demonstrate that purifying selection masking an ancient evolutionary history is not a phenomenon constrained to negative-sense RNA viruses but can be seen in positive-sense RNA viruses like coronaviruses as well.
Interestingly, our extrapolated estimate of the tMRCA of coronaviruses infecting mammalian (alphacoronaviruses and betacoronaviruses) and avian (gammacoronaviruses and deltacoronaviruses) species of 190 to 489 (mean of 293) million years ago corresponds to the inferred tMRCA of these host species based on fossil and molecular evidence of around 325 million years ago. It is tempting to speculate that this correspondence between dates is evidence of coevolution and codivergence between bat and avian species and the coronavirus genera. However, given the uncertainty associated with branch length estimation under strong selection and the extreme saturation leading to a dramatic increase in the inferred tree length, our analyses may not provide an accurate estimation of the tMRCA of coronaviruses. Nevertheless, our results strongly suggest that the 10,000-year-ago tMRCA of coronaviruses is underestimated by orders of magnitude. These results leave open the possibility that coronaviruses have been infecting bats and/or birds since the origin of these clades tens of millions of years ago or possibly since their divergence from each other in the carboniferous period, over 300 million years ago. This extrapolation, rather than be considered a reliable estimate of the coronavirus tMRCA, should be viewed as a biologically plausible hypothesis based on realistic parameters (e.g., patterns of substitution rates and selection profiles). We can no longer reject an ancient coevolutionary relationship based on the molecular clock.
The degree to which host switching and codivergence have shaped coronavirus diversity remains unresolved. The observation of recent viral cross-species transmission events among mammalian and avian species is clear evidence of recent host switching. Conversely, the separation of mammalian and avian coronaviruses into distinct genera is suggestive of codivergence: mammalian coronaviruses (i.e., alphacoronaviruses and betacoronaviruses) are generally inferred to be reciprocally monophyletic. Therefore, a formal analysis of host switching and codivergence in coronaviruses would be useful for disentangling which sections of the coronavirus phylogeny represent codivergence and which represent host-switching events.
For the shorter branches (≤0.05 substitution per site) in the coronavirus phylogenetic trees, we found general agreement in the inferred branch lengths between evolutionary models (GTR + Γ4 and BS-REL); there was no evidence of underestimation of the lengths of short branches. Therefore, for closely related viral lineages involved in recent zoonotic transmission events (e.g., SARS coronavirus in humans, bats, and civet cats), previous dating estimates are consistent with our findings. Furthermore, this observation suggests that substitution rate estimates inferred from these recent outbreaks are robust to the biasing effects of purifying selection with respect to branch length estimation (though coalescent effects may still have an impact on these recent evolutionary rate estimates). However, it remains unclear how the lower-than-average mutation rate of coronaviruses (10−5 to 10−6 mutation per site per replication) translates into a typical short-term substitution rate (10−3 substitution per site per year). Further investigation on this topic is needed.
BS-REL is an attractive tool for future studies of ancient virus evolution. The use of evolutionary models that allow for variable selection pressure across all branches in a phylogeny when estimating branch lengths is an advance over previous implementations by Wertheim and Kosakovsky Pond, which necessitate a priori identification of long internal branches bearing the mark of strong purifying selection. Unlike the phylogenetic trees in this previous study, which were characterized by closely related isolates separated by long internal branches, the coronavirus phylogenies are complicated, with a mixture of long and short branches interspersed throughout the tree. The BS-REL framework represents a flexible and powerful approach to the modeling of natural selection in the evolution of viruses, and it does not necessitate designating which branches experienced stronger or weaker selection pressures.
Moreover, like our previous approach to the analysis of Ebola and avian influenza viruses, BS-REL found evidence of branches experiencing saturation. Because of the way in which BS-REL parameterizes variable selection, the saturated branches were estimated to be longer in coronaviruses than in Ebola and avian influenza viruses.
The BS-REL model almost certainly overfits data on short branches with simple patterns of natural selection, which likely affects the accuracy of branch length estimates across the tree. In the case of coronaviruses, this overfitting is not a serious problem because the expansion of branch length estimates due to saturation is extreme and unlikely to produce precise estimates. A more parsimonious implementation of BS-REL would be useful for addressing issues in viral evolution in which more precise branch length estimates are needed. Appropriate modeling of variation in the strength of natural selection will be integral for obtaining more accurate tMRCAs of viral lineages. Furthermore, it is possible that employing more realistic evolutionary models, for example, in maximum-likelihood or Bayesian tree searches, could improve the quality of viral phylogenetic inference.
In summary, our results indicate that coronaviruses have an evolutionary history much longer than those suggested by phylogenetic trees inferred by using standard nucleotide evolution models. This finding allows for a coevolutionary relationship between coronaviruses and their natural hosts. It is possible that such a long-term relationship has allowed some animal species (e.g., bats) to evolve strategies to coexist with coronaviruses and vice versa. Further investigation of this topic might help us to better understand virus-host coevolution, the origin of coronaviruses, and other related viral families in the order Nidovirales.
—————————————————————————
—————————————————————————
Ancient Mystery Viruses Discovered Sealed Within Tibetan Glacier For 15,000 Years
2020
https://in.mashable.com/science/10763/ancient-mystery-viruses-discovered-sealed-within-tibetan-glacier-for-15000-years
—————————————————————————
Coronaviruses in Avian Species – Review with Focus on Epidemiology and Diagnosis in Wild Birds
2018
https://www.researchgate.net/figure/List-of-all-identified-coronavirus-species-within-individual-genera-according-to-virus_tbl1_328593345

(List of all identified coronavirus species within individual genera according to virus taxonomy released in 2016).

Summary of monitoring studies on coronavirus prevalence in wild birds
————————————————————————-
————————————————————————-
Alphacoronaviruses Detected in French Bats Are Phylogeographically Linked to Coronaviruses of European Bats
2015

List of sequences used for phylogeny analyses with Genbank accession number, coronavirus group, host species and geographic origin and name used in this study.
————————————————————————-
————————————————————————-
Scientists Discover 6 New Coronaviruses In Bats
May 19, 2020
https://www.youtube.com/watch?v=VXDWFkKhiqg
————————————————————————-
Coronaviridae and SARS-associated Coronavirus Strain HSR1
2004
Abstract
During the recent severe acute respiratory (SARS) outbreak, the etiologic agent was identified as a new coronavirus (CoV). We have isolated a SARS-associated CoV (SARS-CoV) strain by injecting Vero cells with a sputum specimen from an Italian patient affected by a severe pneumonia; the patient traveled from Vietnam to Italy in March 2003. Ultrastructural analysis of infected Vero cells showed the virions within cell vesicles and around the cell membrane. The full-length viral genome sequence was similar to those derived from the Hong-Kong Hotel M isolate. By using both real-time reverse transcription–polymerase chain reaction TaqMan assay and an infectivity plaque assay, we determined that approximately 360 viral genomes were required to generate a PFU. In addition, heparin (100 μg/mL) inhibited infection of Vero cells by 50%. Overall, the molecular and biologic characteristics of the strain HSR1 provide evidence that SARS-CoV forms a fourth genetic coronavirus group with distinct genomic and biologic features.An outbreak of atypical pneumonia, referred to as severe acute respiratory syndrome (SARS), was identified in the Guangdong Province of People’s Republic of China at the end of 2002 and spread to other Asian countries and Canada from February through March 2003. Individual cases (all in persons infected in Asia) were diagnosed in Europe during the same period. A novel human coronavirus (SARS-associated coronavirus [SARS-CoV]) has been isolated from the oropharyngeal specimens of patients with SARS. Experimental infection of macaques has confirmed that the SARS-CoV is the cause of SARS.
Coronaviruses are enveloped, positive-stranded RNA viruses associated with enteric and respiratory diseases in animals and humans; they are currently classified into three antigenic groups: group 1 and 2 include mammalian coronaviruses, and group 3 encompasses avian coronaviruses. Human coronaviruses are associated with common cold-like diseases and are included in both group 1 (CoV-229E) and 2 (CoV-OC43). Sequence analysis of the complete genome of SARS-CoV has shown an RNA molecule of about 29,750 bases in length, with a genome organization similar to that of other coronaviruses. In spite of this similar organization, the SARS-CoV RNA sequence is only distantly related to that of previously characterized coronaviruses. Consequently, whether the SARS-CoV has “jumped” from a nonhuman host reservoir to humans and the molecular basis of such a jump remain unanswered questions. Some biologic features of the SARS-CoV described in vivo and in vitro differ from those of other coronaviruses previously identified. Among these features are the peculiar tropism of the virus for Vero cells (a continuous cell line established from monkey kidney epithelial cells), its capacity for growth at 37°C (while other respiratory coronaviruses grow at lower temperatures), and its ability to infect lower respiratory tract tissues. These aspects render the molecular and biologic characterization of SARS-CoV important not only for understanding the determinants of its pathogenic potential but also for planning rational strategies of antiviral therapy and vaccination.
We have recently obtained a SARS-CoV isolate from a frozen sputum sample collected from an Italian patient affected by a respiratory disease of unknown cause; onset of illness began during the patient’s travel to Vietnam in March 2003. The viral strain has been designated as SARS-CoV HSR1. To gain insight into SARS-CoV biopathology, we analyzed the relevant features of SARS-CoV HSR1 growth in vitro, including the ultrastructural analysis of the consequences of virus replication in Vero cells. We have optimized both a reverse transcription–polymerase chain reaction (RT-PCR) TaqMan assay for quantifying the number of viral genomes and a plaque assay for performing titration of the virus infectivity. In addition, we have completely sequenced the viral genome of the SARS-CoV HSR1 and compared it to other SARS-CoV strains recently isolated in disease-epidemic areas.
Methods
SARS-CoV Isolation
In March 2003, an Italian man who recently traveled from Vietnam to Italy was affected by a respiratory disease of unknown cause; he was hospitalized in a clinical unit for acute infectious diseases in a public hospital in Milan, Italy. A sputum sample was collected at the peak of illness and stored at –80°C. Isolation of SARS-CoV was performed with Vero cells maintained in Dulbecco’s modified Eagle medium (D-MEM, BioWhittaker, Verviers, Belgium) and supplemented with 10% fetal calf serum (FCS, HyClone, Perbio Science Erembodegem-Aalst, Belgium), penicillin/streptomycin (BioWhittaker), and 2.5 μg/mL Fungizone (Invitrogen Ltd, Life Technologies, Paisley, UK) (complete medium). In detail, an aliquot (0.5 mL) of the sputum sample was mixed with 2 x 106 Vero cell suspension in 2 mL of complete medium. After incubation for 1 h at 37°C, 5% CO2, 4 mL of complete medium was added, and the cell suspension was transferred into a 25-cm2 tissue culture flask (Falcon, Becton Dickinson Labware, Lincoln Park, NJ). Twenty-four hours after inoculation, the cell cultures were examined with an optical microscope for evidence of cytopathic effects (CPE). An aliquot of culture supernatant was collected and stored at –80°C for RT-PCR evaluation. After an additional 24 hours, both the culture supernatant and the cells were collected after 2 cycles of freeze thawing followed by clarification of the thawed contents by centrifugation at 1,000 x g and dispensation of the supernatant into aliquots stored at –80°C (primary viral stock). For generation of the secondary and tertiary viral stocks, adherent Vero cells flasks were seeded in a 25 cm2 tissue culture and injected with 0.5 mL of stored supernatant filtered with 0.45 μ filters. Three days after infection, the cells and supernatant were subjected to 2 cycles of freeze thawing, and the supernatant was collected after centrifugation followed by filtration as described above (secondary viral stock). Finally, Vero cells seeded in 2 flasks of 75 cm2 were injected with 1.5 mL of the second passage virus stock in a total volume of 25 mL to generate a third-passage viral stock.
RT-PCR Amplification of Viral Sequences
Direct SARS-CoV RNA amplification was performed starting from the sputum sample treated with an equal volume of phosphate-buffered solution (PBS). RNA was purified from 750 µL of the resulting homogenate by using the Qiagen Viral RNA Mini Kit (Qiagen, Inc., Santa Clarita, CA) (elution volume 50 μL), according to manufacturer’s instructions. Viral RNA was extracted from 0.5 mL of culture supernatant with the same kit (elution volume 50 μL). cDNA synthesis and subsequent amplification were performed by using either nested RT-PCR or real-time RT-PCR approach, as described elsewhere, with minor modifications (3). In brief, 5 μL of extracted RNA was reverse-transcribed for 30 min at 42°C by using Mo-MuLV RT, a mixture of random hexamers, and an oligo-(dT) primer. Two microliters of the synthesized cDNA was subsequently amplified by using the following primers pairs: outer primers: BNI-OUTS2, 5′-ATG AAT TAC CAA GTC AAT GGT TAC-3′; BNI-OUTAS, 5′-CAT AAC CAG TCG GTA CAG CTA C-3′; inner primers: BNI-INS, 5′-GAA GCT ATT CGT CAC GTT CG,-3′; BNIINAS, 5′-CTG TAG AAA ATC CTA GCT GGA G-3′. A sequence of the SARS-CoV RNA encompassing the target region was used as positive control, and a sputum sample from a SARS-negative healthy donor and a feline coronavirus RNA extract were used as negative controls. The amplified PCR product was separated by agarose gel electrophoresis visualized by ethidium bromide staining and sequenced directly on an ABI PRISM 3100 Genetic Analyzer with ABI PRISM BigDye Terminator v3.0 sequencing kit (Applied Biosystems, Foster City, CA). SARS-CoV was then identified by searching for homologies between the amplified fragment and previously deposited sequences using BLAST. Finally, the quantitation of the viral RNA was performed in a TaqMan assay after generation of cDNA. The primer pair and probe BNITMSARS1, 5′-TTATCACCCGCGAAGAAGCT-3′; BNITMSARAS2, 5′-CTCTAGTTGCATGACAGCCCTC-3′, BNI-TMSARP 6-carboxifluorescein-TCG TGC GTG GAT TGG CTT TGA TGT-6 carboxy-tetramethylrhodamin (3) were added to the universal PCR master mix (Applied Biosystems) at 200 and 120 nM, respectively, in a final volume of 25 μL. The standard was obtained by cloning the 77-bp fragment into the pCR2.1 plasmid by using the TA cloning kit (InVitrogen Corp., San Diego, CA). A linear distribution (r = 0.99) was obtained between 101 to 108 copies.
SARS-CoV Plaque Infectivity Assay
To have the most effective quantitative assay of virus infectivity and to compare the infectious titer with quantitative molecular assays, we optimized a plaque assay (determining the PFU/mL). Confluent Vero cells in 6 well plates (Falcon) were incubated in duplicate with 1 mL of PBS containing 100 μL of SARS-CoV HSR1 viral stock in 10-fold serial dilutions from –10e2 (1/100) to –10e8 (1/100,000,000). After 1 hour of incubation, the viral inoculum was removed and 1 mL of 1% carboxymethylcellulose (Sigma Chemical Corp., St. Louis, MO) overlay with DMEM supplemented with 1% fetal calf serum was added to each well. After 6 days of incubation, the cells were stained with 1% crystal violet (Sigma) in 70% methanol. The plaques were counted after being examined with a stereoscopic microscope (SMZ-1500, Nikon). The virus titer was calculated in PFU per milliliter.
Ultrastructural Analysis
Adherent Vero cells were infected with 5 x 104 PFU/mL of SARS-CoV HSR1 at the third passage in a T-75 flask. Twenty-four hours after infection, Vero cells were detached from the tissue culture flask with a cell scraper, and the cell suspension was centrifuged at 173 x g for 4 min. The pellet was resuspended in PBS without Ca++ and Mg++, and the suspension was spun at 173 x g for an additional 4 min. The cell pellet was resuspended and fixed in 4% formaldehyde and 2.5% glutaraldehyde in cacodylate buffer and incubated for 5 min at room temperature. The sample was then centrifuged at 13,414 x g for 5 min; the pellet was fixed with 2% OsO4 in 2.5% glutaraldehyde in cacodylate buffer for 60 minutes. The pellet was dehydrated in graded ethanol, washed in propylene oxide and infiltrated for 12 hours in a 1:1 mixture of propylene oxide and epoxidic resin (Epon). Cells were then embedded in Epon and polymerized for 24 hours at 60°C. Slides were cut with ultramicrotome (Ultracut Uct, Leica, Deerfield, IL), stained with uranyl acetate and lead citrate, and metaled. The ultrathin sections of infected Vero cells were observed through transmission electron microscopy (Hitachi H7000).
SARS-CoV HSR1 Genome RNA and Phylogenetic Reconstruction
Sequencing of the complete SARS-CoV HSR1 genome was performed by using 68 partially overlapping primers encompassing the whole viral genome. A 5′ rapid amplification of PCR ends (RACE) technique was performed to capture the 5′-untranslated region of the genome (10). Each 750-bp fragment was gel-isolated by means of a QIAQuick gel Extraction kit (Qiagen) and directly sequenced from both directions inward and outward. SeqScape version 2.0 (Applied Biosystems) software was used for base calling, editing, and assembly of the fragments. Manual check of the differences between the electropherograms and the reference sequence (Urbani isolate) was performed and eventually led to reamplification or resequencing of some fragments. The complete sequence of SARS-CoV HSR1 strain was aligned with all the previously sequenced full-length genomes available from the GenBank database by using ClustalW and editing with BioEdit version 5.0.9 for manual corrections. Available full-length sequences of SARS-CoV strains accessed from GenBank and used for phylogenetic analysis and genotyping were as follows: Taiwan TC2 (AY338175), Taiwan TC1 (AY338174), TWC (AY321118), Sin2774 (AY283798), Sin2748 (AY283797), Sin2679 (AY283796), Sin2677 (AY283795), Sin2500 (AY283794), Frankfurt 1 (AY291315), BJ04 (AY279354), BJ03 (AY278490), BJ02 (AY278487), GZ01 (AY278489), CUHK-W1 (AY278554), ZJ01 (AY297028), TOR2 (AY274119), TW1 (AY291451), CUHKSu10 (AY282752), BJ01 (AY278488), Urbani (AY278741), and HKU-39849 (AY278491). Sequences were trimmed to equivalent length and phylogenetic relationships were estimated with PAUP* (maximum-parsimony and maximum-likelihood methods by using the p-distance model) and MEGA version 2.1. Trees were edited by using Treeview.
Results
Figure 1. Cytopathic effect (CPE) of primary severe acute respiratory syndrome–associated coronavirus strain HSR1 isolate. A, uninfected Vero cells form a continuous monolayer of spindle-shaped cells. B, a strong CPE was observed after…
Both nested RT-PCR and the real-time RT-PCR assays performed on the sputum sample from a person with SARS tested positive for SARS-CoV RNA. The viral load in the sample was estimated to be 5.6 x 104 SARS-CoV RNA copies/mL. Twenty-four hours after injection of Vero cells with the sputum sample, a strong CPE was observed, as indicated in Figure 1B. The CPE was diffused with cell rounding with refractive appearance, and the cell monolayer was destroyed as compared to control uninfected Vero cells (Figure 1A). The viral load in the culture supernatant 24 hours after injection with the clinical sample was 1.3 x 105 copies/mL as determined by quantitative real-time RT-PCR. Serial passage of the virus on Vero cells to obtain a tertiary viral stock yielded 9.1 x 108 copies/mL. A plaque assay was optimized to determine the in vitro infectivity of SARS-CoV. The tertiary viral stock tested 2.5 x 106 PFU/mL in the plaque assay, suggesting that about 360 genomes were required to generate a single plaque in tissue cultures, at least under the conditions described here. We also used the plaque assay for testing the potential inhibitory effect on virus infectivity of a single concentration of heparin, the prototypic compound of a class of inhibitors of virus entry for enveloped viruses including HIV type 1 (HIV-1) and herpes simplex virus 1 and 2 (HSV-1 and 2). Heparin indeed reduced the formation of plaques by 50% when added 30 min before infection of Vero cells with 100 PFU/mL of the SARS-CoV (Figure 2).
Figure 3. Ultrastructural analysis of Vero cells infected with severe acute respiratory syndrome–associated coronavirus (SARS-CoV) strain HSR1. A, intracellular budding of SARS-CoV in large vesicles containing CoV virions (magnification x30,000); B, clusters of… Thin-section electron microscopy showed the typical features of intracellular CoV particles. Cells were engulfed with viral particles localized in cytoplasmic vesicles (Figure 3A). This feature is typical of all Coronaviridae viruses that bud intracellularly at membranes of the intermediate compartment between the endoplasmic reticulum and the Golgi complex, whereas newly assembled virions reach the cell surface by vesicular transport. After the extracellular release, virus particles were found in large clusters adjacent to the plasma membrane, as evidenced in Figure 3B. Overall, the ultrastructural analysis documented that SARS-CoV cultivated in Vero cells behaved like a typical coronavirus, characterized by intracellular budding.
Figure 4. Phylogenetic tree obtained by applying PAUP* (maximum-likelihood methods using the p-distance model) applied to complete genome sequences of the severe acute respiratory syndrome–associated coronavirus (SARS-CoV) HSR1 strain and the 21 other…
The sequence of SARS-CoV strain HSR1 genomic RNA was 29,751 bases in length, with a polyanion tail. We identified the major open reading frames (ORFs), coding for the 4 major structural proteins, namely: spike (S), envelope (E) matrix (M), nucleocapsid (N) gene products, and at least other 10 proteins, including a few of unknown function. The complete sequence of SARS-CoV strain HSR1 has been deposited in the GenBank database (GenBank accession no. AY323977). This sequence was aligned with those of the other 21 SARS-CoV isolates to facilitate phylogenetic analysis. Overall, the mean difference in nucleotide composition between all the isolates was 18 ± 1 nt variations within the whole genomes, thus confirming the genetic conservation observed previously (9). On the whole, 149 sites were variable among the 22 aligned isolates, and 24 loci in the viral genome varied in more than one isolate, including in the SARS-CoV HSR1 genome (recurrent mutations). Eleven of 24 recurrent mutations were silent, whereas 3 of 13 mutations generating amino acid substitutions were observed within the N-terminal domain of the spike glycoprotein gene (positions 21,722, 22,223, and 22,423, determining a G to D, an I to T, and a G to R amino acid change, respectively). Six mutations were detected in the replicase gene (positions 8,572, 9,404, 9,479, 9,854, 17,564, and 19,084, determining a V to L, a V to A, a V to A, an A to V, a D to E, and a T to I amino acid change, respectively), and 1 mutation was found in the matrix protein (position 26,600, determining an A to V amino acid change). The phylogenetic analysis was also performed by using the maximum parsimony method, considering only sequence variants that recurred in more than one strain in order to reduce the mutational noise caused by PCR or sequencing mistakes. Figure 4 shows the phylogenetic tree obtained with the maximum likelihood method. The maximum parsimony trees produced the same structure with minor changes in the subtrees’ branch patterns (data not shown). In these analyses, SARS-CoV HSR1 appears to be strongly related to strains isolated from patients who had traveled from Hong Kong to different geographic areas (Singapore, Canada, Vietnam) and spread the infection to their home countries in a few cases.
————————————————————————–
Could Intravenous Immunoglobulin Collected from Recovered Coronavirus Patients Protect against COVID-19 and Strengthen the Immune System of New Patients?
2020 Mar
https://www.ncbi.nlm.nih.gov/pubmed/32218340
Abstract
The emergence of the novel coronavirus in Wuhan, China, which causes severe respiratory tract infections in humans (COVID-19), has become a global health concern. Most coronaviruses infect animals but can evolve into strains that cross the species barrier and infect humans. At the present, there is no single specific vaccine or efficient antiviral therapy against COVID-19. Recently, we showed that intravenous immunoglobulin (IVIg) treatment reduces inflammation of intestinal epithelial cells and eliminates overgrowth of the opportunistic human fungal pathogen Candida albicans in the murine gut. Immunotherapy with IVIg could be employed to neutralize COVID-19. However, the efficacy of IVIg would be better if the immune IgG antibodies were collected from patients who have recovered from COVID-19 in the same city, or the surrounding area, in order to increase the chance of neutralizing the virus. These immune IgG antibodies will be specific against COVID-19 by boosting the immune response in newly infected patients. Different procedures may be used to remove or inactivate any possible pathogens from the plasma of recovered coronavirus patient derived immune IgG, including solvent/detergent, 60 °C heat-treatment, and nanofiltration. Overall, immunotherapy with immune IgG antibodies combined with antiviral drugs may be an alternative treatment against COVID-19 until stronger options such as vaccines are available.
—————————
Deadly Black Fungus in COVID Patients – Mucormycosis
Jun 2, 2021
https://www.youtube.com/watch?v=bUtfSmSJbM8
—————————
How a deadly fungal infection shape-shifts into an invasive monster
November 20, 2018
Summary:
Researchers have shed new light on just how the fungal infection Candida albicans shape-shifts into a deadly version with hyphae or filaments that help it break through human tissues and into the bloodstream. Understanding this process is key to the development of drugs against this fatal infection.
https://www.sciencedaily.com/releases/2018/11/181120125928.htm
—————————
Fluconazole makes fungi sexually active
Date:
February 8, 2019
Summary:
Under the influence of the drug fluconazole, the fungus Candida albicans can change its mode of reproduction and thus become even more resistant, scientists report.
https://www.sciencedaily.com/releases/2019/02/190208115329.htm
——————————–
Growth, Morphogenesis, and Virulence of Candida albicans after Oral Inoculation in the Germ-Free and Conventional Chick
1966
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC316115/
Abstract
Balish, Edward (Syracuse University, Syracuse, N.Y.), and A. W. Phillips. Growth, morphogenesis, and virulence of Candida albicans after oral inoculation in the germ-free and conventional chick. J. Bacteriol. 91:1736–1743. 1966.—The effects of intestinal bacteria on the multiplication, morphogenesis, and infectivity of Candida albicans in the alimentary tract were investigated by comparing results obtained in germ-free and conventional chicks after oral inoculation. This challenge resulted in the establishment of large numbers of the pathogen in the alimentary tract of each group of chicks; these numbers were increased in crop contents from challenged bacteria-free chicks wherein hyphae predominated over the yeast form. These animals also had lesions of the crop epithelium containing numerous hyphae and few yeast-like forms. In contrast, challenged conventional chicks receiving an adequate diet displayed no evidence of infection. Their alimentary tract contained the yeast form of C. albicans; no hyphae were seen. Although we found bacterial inhibition of C. albicans multiplication in the alimentary tract, this in itself did not seem to explain the resistance to intestinal candidiasis in our conventional chicks. We argued that this resistance to infection was due chiefly to the prevention of hyphal development in C. albicans by intestinal bacteria. C. albicans in the gut of our conventional chicks resulted in some increase in numbers of enterococci in contents from the crop. Increased pH values in contents from the gut of germ-free chicks were not clearly related to infection after challenge. The Eh of the above crop contents were only slightly decreased in the germ-free crop. Thus the Eh did not appear to be involved in susceptibility to infection. Invasion of the blood stream and kidneys of conventional chicks by the yeast form of C. albicans occurred in challenged animals receiving a purified diet which had been radiation-sterilized and stored for 6 months at room temperature (25 C). Their growth rate decreased and they became moribund; no hyphae were observed in tissues or intestine of these animals. Challenged bacteria-free chicks receiving the same diet were resistant to the above invasion, although they had crop lesions containing hyphae as described. The resistance of these chicks to systemic invasion was attributed to absence of intestinal bacteria competing for low levels of vitamins in the stored diet. Germ-free chicks had decreased levels of serum γ-globulin which increased after challenge, whereas this value was unchanged in conventional birds after challenge.
——————————-
Candida auris Infection in COVID-19 Patients
November 6, 2020
Patients with COVID-19 hospitalized in intensive care in India developed multidrug-resistant C. auris candidemia associated with a 60% mortality rate.
The COVID-19 pandemic is emerging or reemerging in many countries worldwide and in nearly all U.S. states. Intensive care units (ICUs) are at capacity, and trained ICU staff are in short supply. The most susceptible patients are older and have comorbid conditions, and secondary infections in this population have been reported increasingly. These researchers describe 15 cases of nosocomial candidemia among 2.5% of 596 patients with documented COVID-19 admitted to a New Delhi, India, ICU from April through July 2020. Of these cases, 10 (67%) had Candida auris isolated from their bloodstream. Two of these 10 also had C. auris in their urine. C. albicans was the next most common fungus (3 cases); C. tropicalis and C.krusei each infected one patient. C. auris developed 10 to 42 days after hospital admission. All of these patients had prolonged hospitalization, chronic comorbidities, and indwelling catheters, and half received mechanical ventilation. Overall mortality in the 15 cases with candidiasis was 53%, but C. auris–related mortality was 60%.
All C. auris isolates were resistant to fluconazole, 30% were nonsusceptible to voriconazole, 40% were resistant to amphotericin B, and 60% were resistant to 5-flucytosine. Overall, 70% were multidrug-resistant. All isolates were sensitive to echinocandins.
https://www.jwatch.org/na52745/2020/11/06/candida-auris-infection-covid-19-patients
——————————-
COVID-19 Impairs Immune Response to Candida albicans
2021
https://pubmed.ncbi.nlm.nih.gov/33717195/
Abstract
Infection with SARS-CoV-2 can lead to Coronavirus disease-2019 (COVID-19) and result in severe acute respiratory distress syndrome (ARDS). Recent reports indicate an increased rate of fungal coinfections during COVID-19. With incomplete understanding of the pathogenesis and without any causative therapy available, secondary infections may be detrimental to the prognosis. We monitored 11 COVID-19 patients with ARDS for their immune phenotype, plasma cytokines, and clinical parameters on the day of ICU admission and on day 4 and day 7 of their ICU stay. Whole blood stimulation assays with lipopolysaccharide (LPS), heat-killed Listeria monocytogenes (HKLM), Aspergillus fumigatus, and Candida albicans were used to mimic secondary infections, and changes in immune phenotype and cytokine release were assessed. COVID-19 patients displayed an immune phenotype characterized by increased HLA-DR+CD38+ and PD-1+ CD4+ and CD8+ T cells, and elevated CD8+CD244+ lymphocytes, compared to healthy controls. Monocyte activation markers and cytokines IL-6, IL-8, TNF, IL-10, and sIL2Rα were elevated, corresponding to monocyte activation syndrome, while IL-1β levels were low. LPS, HKLM and Aspergillus fumigatus antigen stimulation provoked an immune response that did not differ between COVID-19 patients and healthy controls, while COVID-19 patients showed an attenuated monocyte CD80 upregulation and abrogated release of IL-6, TNF, IL-1α, and IL-1β toward Candida albicans. This study adds further detail to the characterization of the immune response in critically ill COVID-19 patients and hints at an increased susceptibility for Candida albicans infection.
——————————-
COVID Unleashes the ‘Lurking Scourge’ Candida Auris
November 11, 2020
https://www.infectioncontroltoday.com/view/covid-unleashes-the-lurking-scourge-candida-auris
——————————-
Candida auris Outbreak in a COVID-19 Specialty Care Unit — Florida, July–August 2020
On January 8, 2021, this report was posted online as an MMWR Early Release.
https://www.cdc.gov/mmwr/volumes/70/wr/mm7002e3.htm
——————————-
Staphylococcus aureus colonization and non-influenza respiratory viruses: Interactions and synergism mechanisms
2018
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6177244/
ABSTRACT
Viral infections of the respiratory tract can be complicated by bacterial superinfection, resulting in a significantly longer duration of illness and even a fatal outcome. In this review, we focused on interactions between S. aureus and non-influenza viruses. Clinical data evidenced that rhinovirus infection may increase the S. aureus carriage load in humans and its spread. In children, respiratory syncytial virus infection is associated with S. aureus carriage. The mechanisms by which some non-influenza respiratory viruses predispose host cells to S. aureus superinfection can be summarized in three categories: i) modifying expression levels of cellular patterns involved in S. aureus adhesion and/or internalization, ii) inducing S. aureus invasion of epithelial cells due to the disruption of tight junctions, and iii) decreasing S. aureus clearance by altering the immune response. The comprehension of pathways involved in S. aureus-respiratory virus interactions may help developing new strategies of preventive and curative therapy.
Staphylococcus aureus and other respiratory tract viruses
S. aureus interactions with rhinovirus or RSV have been actively, yet insufficiently, investigated. However, bacterial interactions with other respiratory viruses are poorly studied and only partial studies are available. For example, parainfluenza virus has been shown to enhance the ability of non-typeable H. influenzae and S. pneumoniae to adhere to human respiratory epithelial cells. However, there is no information available about the interaction of these agents with S. aureus in humans. Only one in vitro study conducted on bovine embryonic lung cells investigated the effect of bovine parainfluenza virus infection on adherence of several bacterial agents, including S. aureus, and found no virus-specific effect on any of the tested bacteria. Besides, normal human bronchial epithelial cells that were pre-incubated with S. pneumoniae resulted in an increased susceptibility to infection with human metapneumovirus. Nevertheless, this was not the case for cells pre-incubated with H. influenzae, M. catarrhalis or S. aureus. On the contrary, synergistic effect between S. aureus and coronavirus has been demonstrated in vivo in a swine model. Lipoteichoic acid from S. aureus increased the susceptibility to coronavirus infection in pigs via increased secretion of pro-inflammatory cytokines IL-6, IL-12, IL-23 and IFN-γ. To date, the lack of clinical and experimental data about the relationship between S. aureus colonization and other respiratory viruses complicates the understanding of potential interactions between these pathogens.
—————————————————————————-
Infection with human coronavirus NL63 enhances streptococcal adherence to epithelial cells
2011 Jun
Abstract
Understanding the mechanisms of augmented bacterial pathogenicity in post-viral infections is the first step in the development of an effective therapy. This study assessed the effect of human coronavirus NL63 (HCoV-NL63) on the adherence of bacterial pathogens associated with respiratory tract illnesses. It was shown that HCoV-NL63 infection resulted in an increased adherence of Streptococcus pneumoniae to virus-infected cell lines and fully differentiated primary human airway epithelium cultures. The enhanced binding of bacteria correlated with an increased expression level of the platelet-activating factor receptor (PAF-R), but detailed evaluation of the bacterium–PAF-R interaction revealed a limited relevance of this process.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3168281/

Adhesion of Streptococcus pneumoniae to LLC-MK2 cells (a) and HAE cultures (b) is increased following infection with HCoV-NL63. Fluorescence images from FITC-labelled bacteria and bright-field images were obtained using a fluorescence microscope. Magnification: ×200 (a); ×100 (b). Data are representative of two independent experiments.
————————————————————————–
Staphylococcus aureus Bacteremia in Patients Infected With COVID-19: A Case Series
Nov 2020
https://pubmed.ncbi.nlm.nih.gov/33269299/
————————————————————————–
Staphylococcus aureus ventilator-associated pneumonia in patients with COVID-19: clinical features and potential inference with lung dysbiosis
June 2021
https://pubmed.ncbi.nlm.nih.gov/34099016/
————————————————————————–
Proven Fatal Invasive Aspergillosis in a Patient with COVID-19 and Staphylococcus aureus Pneumonia
Mar 2021
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8003593/
————————————————————————–
A Staphylococcus aureus Coinfection on a COVID-19 Pneumonia in a Breast Cancer Patient
2020
https://pubmed.ncbi.nlm.nih.gov/33061542/
————————————————————————–
Panton-Valentine Leukocidin–Secreting Staphylococcus aureus Pneumonia Complicating COVID-19
April 16, 2020
https://wwwnc.cdc.gov/eid/article/26/8/20-1413_article
————————————————————————–
Trial of existing antibiotic for treating Staphylococcus aureus bacteremia begins
NIH-supported trial will test dalbavancin in hospitalized adults.
April 27, 2021
https://www.nih.gov/news-events/news-releases/trial-existing-antibiotic-treating-staphylococcus-aureus-bacteremia-begins
————————————————————————–
Covid-19 May Worsen the Antibiotic Resistance Crisis
04.23.2020
The disease can’t be treated with these drugs, but antibiotic use is rising anyway, in ICUs and among the worried well.
https://www.wired.com/story/covid-19-may-worsen-the-antibiotic-resistance-crisis/
————————————————————————–
Taking Aim at Antibiotic-Resistant Bacteria During COVID
2020
https://www.infectioncontroltoday.com/view/taking-aim-at-antibiotic-resistant-bacteria-during-covid
————————————————————————–
Antibiotic resistance: the hidden threat lurking behind Covid-19
March 23, 2020
https://www.statnews.com/2020/03/23/antibiotic-resistance-hidden-threat-lurking-behind-covid-19/
————————————————————————–
Scientists discover antibodies that may neutralize a range of SARS-CoV-2 variants
September 6, 2021
https://www.medicalnewstoday.com/articles/scientists-discover-antibodies-that-may-neutralize-a-range-of-sars-cov-2-variants
————————————————————————–
Bispecific antibodies: a mechanistic review of the pipeline
2019 Aug
https://pubmed.ncbi.nlm.nih.gov/31175342/
Abstract
The term bispecific antibody (bsAb) is used to describe a large family of molecules designed to recognize two different epitopes or antigens. BsAbs come in many formats, ranging from relatively small proteins, merely consisting of two linked antigen-binding fragments, to large immunoglobulin G (IgG)-like molecules with additional domains attached. An attractive bsAb feature is their potential for novel functionalities – that is, activities that do not exist in mixtures of the parental or reference antibodies. In these so-called obligate bsAbs, the physical linkage of the two binding specificities creates a dependency that can be temporal, with binding events occurring sequentially, or spatial, with binding events occurring simultaneously, such as in linking an effector to a target cell. To date, more than 20 different commercialized technology platforms are available for bsAb creation and development, 2 bsAbs are marketed and over 85 are in clinical development. Here, we review the current bsAb landscape from a mechanistic perspective, including a comprehensive overview of the pipeline.
————————————————————————–
Bispecifics: The Next Weapon Against Cancer
2021
https://www.statnews.com/events/bispecifics-the-next-weapon-against-cancer/
————————————————————————–
Antibiotic approved for skin infections; COVID-linked rise in hospital infections; School policy on antibiotics in turkey
2021
https://www.cidrap.umn.edu/news-perspective/2021/03/stewardship-resistance-scan-mar-16-2021
————————————————————————–
Too Many COVID-19 Patients Get Unneeded “Just in Case” Antibiotics
August 27, 2020
Study in 38 Michigan hospitals shows need for faster in-hospital testing for coronavirus and other infections, which could reduce overuse of antibiotics.
https://labblog.uofmhealth.org/rounds/too-many-covid-19-patients-get-unneeded-just-case-antibiotics
————————————————————————–
Can antibiotics treat COVID-19 (coronavirus)? And other treatments
June 24, 2020
https://www.medicalnewstoday.com/articles/can-antibiotics-treat-the-coronavirus-disease
————————————————————————–
Episode #11 – Antibiotics & COVID-19
6 November 2020
https://www.who.int/emergencies/diseases/novel-coronavirus-2019/media-resources/science-in-5/episode-11—antibiotics-covid-19
————————————————————————–
These Antibiotics May Offer First-line Treatment For COVID-19
February 27, 2020
https://www.india.com/lifestyle/these-antibiotics-may-offer-first-line-treatment-for-covid-19-3955560/
————————————————————————–
What antibiotics kill Covid-19 (coronavirus)?
Dec 1, 2021
https://www.drugs.com/medical-answers/antibiotics-kill-coronavirus-3534867/
————————————————————————–
Defining variant-resistant epitopes targeted by SARS-CoV-2 antibodies: A global consortium study
23 Sep 2021
Abstract
Antibody-based therapeutics and vaccines are essential to combat COVID-19 morbidity and mortality following severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) infection. Multiple mutations in SARS-CoV-2 that could impair antibody defenses propagated in human-to-human transmission and spillover/spillback events between humans and animals. To develop prevention and therapeutic strategies, we formed an international consortium to map the epitope landscape on the SARS-CoV-2 Spike, defining and structurally illustrating seven receptor-binding domain (RBD)-directed antibody communities with distinct footprints and competition profiles. Pseudovirion-based neutralization assays reveal Spike mutations, individually and clustered together in variants, that impact antibody function among the communities. Key classes of RBD-targeted antibodies maintain neutralization activity against these emerging SARS-CoV-2 variants. These results provide a framework for selecting antibody treatment cocktails and understanding how viral variants might affect antibody therapeutic efficacy.
https://www.science.org/doi/10.1126/science.abh2315?_ga=2.99325726.1613829897.1632143521-2006365807.1575607746
————————————————————————–
Two common drugs could be deadly combination for seniors
February 2, 2015
The combination of a widely used heart medication and a commonly prescribed antibiotic seems to more than double the risk of sudden death in seniors, a new study says.
https://www.cbsnews.com/news/two-common-drugs-could-be-deadly-combination-for-seniors/
————————————————————————–
Epitopes of SARS-CoV-2’s Spike Mapped by International Consortium
September 24, 2021
https://www.genengnews.com/news/epitopes-of-sars-cov-2s-spike-mapped-by-international-consortium/
The Coronavirus Immunotherapy Consortium (CoVIC), an academic-industry, non-profit collaborative research effort, has analyzed antibodies to SARS-CoV-2 that may offer an advantage over traditional antibodies. Originally discovered by Twist Bioscience, the antibodies were found to have potent effects on multiple SARS-CoV-2 variants.
One antibody candidate TB202-3 (CoVIC-094), demonstrated binding to diverse SARS-CoV-2 variant mutations, including strains with the E484K, N501Y, D614G, Y453F, and K417N mutations in pseudovirus assays, indicating this therapeutic antibody may be effective in treating many strains of COVID-19.
The results are published in Science, in the article, “Defining variant-resistant epitopes targeted by SARS-CoV-2 antibodies: A global consortium study.”
“CoVIC analyzed over 250 therapeutic antibodies from 46 groups, in an effort to identify the most effective treatment approaches for patients with COVID-19,” said Erica Ollmann Saphire, PhD, director of CoVIC and professor at the La Jolla Institute for Immunology. “TB202-3 binds to a specific area (RBD-4) of the SARS-CoV-2 spike protein that has not been impacted by most viral mutations. This makes it an important and viable candidate for clinical testing in combination with other antibody therapeutics.”
CoVIC used high-throughput surface plasmon resonance analysis and cryo-EM structural determination, sorting antibodies that react within the SARS-CoV-2 receptor-binding domain (RBD) into 7 different “communities” (RBD-1 through RBD-7). Antibodies in the RBD-4 community bind to the outer face of the RBD and can do so in either the “up” or “down” RBD conformation. Monoclonal antibodies that target RBD-4 bind towards the outer edge of the receptor-binding motif and can block binding to ACE2 on human cells, the entry point for the virus. Select properties of RBD-4 antibodies indicate they may have increased potency against the virus.
COVID-19 continues to evolve, spurring additional mutations and viral strains. TB202-3 binds to a majority of known mutations, with the exception of the L452R mutation present in the Delta and Epsilon variants. Twist developed a new VHH single domain antibody, TB339-031, with a similar structure and potency to TB202-3, which also binds and neutralizes the Delta and Epsilon variants, and that is now advancing through late-stage discovery and validation testing.
“Applying our proprietary drug discovery and optimization platform, we identified and advanced TB202-3 through preclinical studies and submitted this antibody to CoVIC for comprehensive testing in comparison to others,” said Emily Leproust, PhD, CEO and co-founder of Twist Bioscience. “With the continuous emergence of variant strains of SARS-CoV-2, antibodies that bind to regions away from the areas of frequent mutation will be critical to the ongoing global response. While broad-spectrum is a term used most often in relation to antibiotics, it applies here to Twist’s antibodies, as they show efficacy in neutralizing a wide range of SARS-CoV-2 variants.”
“These comprehensive results show that TB202-3 may be more resistant to receptor-driven selection pressure and Twist antibodies may work therapeutically against emerging SARS-CoV-2 variants,” continued Leproust. “In addition to binding location and neutralization efficacy, due to its small size, selectivity, and preclinical efficacy, Twist antibodies may offer an advantage over traditional antibodies as they can be an integral component of a bispecific antibody or can be used with other antibodies. COVID-19 infection continues to spread throughout the world, and new therapeutic options will be required to treat emerging variants.”
Previous studies of TB202-3, a single domain VHH “nanobody,” demonstrated protection against weight loss, a key indicator of disease severity, at the lowest dose of 1 mg/kg in a preclinical hamster challenge model. Immunosuppressed animals were given 1, 5, or 10 mg/kg of each of the Twist antibodies and were assessed for weight loss. Animals treated with all doses of TB202-3 were protected against weight loss, whereas control animals lost a mean of 11.7% of their body weight. Validation and late-stage discovery studies continue for TB339-031.
With VHH-based antibodies able to exhibit pharmaceutically-relevant properties comparable to IgGs, they are a promising therapeutic with several advantages over their bulkier, more complex counterparts. They are also more thermally and chemically stable, making VHH-based therapeutics good candidates to address respiratory infections, administered by inhaler directly to the respiratory tract where the infection is concentrated. In addition, the small size simplifies the manufacturing of VHH antibodies.
————————————————————————–
How antibodies may cause rare blood clots after some COVID-19 vaccines
July 7, 2021
https://www.sciencenews.org/article/coronavirus-covid-vaccine-antibodies-cause-blood-clots-side-effect
————————————————————————–
A Possible Why For The Clotting Problem Of COVID-19 Vaccines
May 29, 2021
https://www.acsh.org/news/2021/05/29/possible-why-clotting-problem-covid-19-vaccines-15572
————————————————————————–
1 of these can end your life (Cipro)
May 15, 2021
https://www.youtube.com/watch?v=joXznzIrB9c
————————————————————————–
Ciprofloxacin
https://en.wikipedia.org/wiki/Ciprofloxacin
Ciprofloxacin is a fluoroquinolone (flor-o-KWIN-o-lone) antibiotic used to treat a number of bacterial infections. This includes bone and joint infections, intra abdominal infections, certain type of infectious diarrhea, respiratory tract infections, skin infections, typhoid fever, and urinary tract infections, among others. For some infections it is used in addition to other antibiotics. It can be taken by mouth, as eye drops, as ear drops, or intravenously.
Common side effects include nausea, vomiting, and diarrhea. Severe side effects include an increased risk of tendon rupture, hallucinations, and nerve damage. In people with myasthenia gravis, there is worsening muscle weakness. Rates of side effects appear to be higher than some groups of antibiotics such as cephalosporins but lower than others such as clindamycin. Studies in other animals raise concerns regarding use in pregnancy. No problems were identified, however, in the children of a small number of women who took the medication. It appears to be safe during breastfeeding. It is a second-generation fluoroquinolone with a broad spectrum of activity that usually results in the death of the bacteria.
Ciprofloxacin was patented in 1980 and introduced in 1987. It is on the World Health Organization’s List of Essential Medicines. The World Health Organization classifies ciprofloxacin as critically important for human medicine. It is available as a generic medication. In 2018, it was the 109th most commonly prescribed medication in the United States, with more than 6 million prescriptions.
————————————————————————–
Guillain-Barre syndrome listed as ‘very rare’ side effect of AstraZeneca vaccine
September 10, 2021
https://nairametrics.com/2021/09/10/guillain-barre-syndrome-listed-as-very-rare-side-effect-of-astrazeneca-vaccine/
————————————————————————–
CDC study shows risk of Guillain-Barré syndrome elevated after Ad.26.COV2.S COVID vaccination
Dec 8 2021
https://www.news-medical.net/news/20211208/CDC-study-shows-risk-of-Guillain-Barre-syndrome-elevated-after-Ad26COV2S-COVID-vaccination.aspx
Background
To date, three vaccines have been authorized for emergency use or FDA-approved in the United States (U.S.) to prevent coronavirus disease 2019 (COVID-19). These vaccines include BNT162b2 (Pfizer-BioNTech – FDA-approved) and mRNA-1273 (Moderna -authorized for emergency use), both of which are mRNA-based vaccines, as well as Ad.26.COV2.S (Janssen -authorized for emergency use).
BNT162b2 and mRNA-1273 are both administered primarily as double-doses, with a third booster shot that has recently been approved after six months. The Ad.26.COV2.S is a replication-incompetent adenoviral vector vaccine and is primarily administered as a single dose; however, the U.S. Food and Drug Administration (FDA) has recently advised Ad.26.COV2.S recipients to receive a booster dose of any approved COVID-19 vaccine after two months.
Guillain-Barré syndrome (GBS) is a rare neurological disorder with an incidence rate of 1-2 per 100,000 person-years. In July 2021, data from the Vaccine Adverse Event Reporting System (VAERS) indicated that more cases of GBS were being reported after vaccination with Ad.26.COV2.S in comparison to other mRNA vaccines.
On July 12, 2021, the U.S. FDA added a warning about GBS to the Ad.26.COV2.S vaccine fact sheet. GBS is being monitored in the Vaccine Safety Datalink as part of ongoing rapid and prospective COVID-19 vaccine safety surveillance efforts.
In a recent study published on the preprint server medRxiv*, researchers compiled available data from the interim analysis of the number of cases of GBS and the risks posed by different vaccines.
————————————————————————–
UK research investigates relationship between COVID-19 vaccination and Guillain-Barré syndrome
Dec 21 2021
https://www.news-medical.net/news/20211221/UK-research-investigates-relationship-between-COVID-19-vaccination-and-Guillain-Barre-syndrome.aspx
————————————————————————–
AstraZeneca’s COVID-19 shot joins the list of vaccines flagged for rare Guillain-Barre syndrome
Sep 9, 2021
https://www.fiercepharma.com/pharma/astrazeneca-s-covid-19-shot-joins-list-vaccines-flagged-for-rare-guillain-barre-syndrome
————————————————————————–
Cyproheptadine
https://en.wikipedia.org/wiki/Cyproheptadine
Cyproheptadine, sold under the brand name Periactin among others, is a first-generation antihistamine with additional anticholinergic, antiserotonergic, and local anesthetic properties.
It was patented in 1959 and came into medical use in 1961.
Medical uses
Periactin (cyproheptadine) 4 mg tablets
Cyproheptadine’s 3D molecular structure represented as Space-filling model
Cyproheptadine is used to treat allergic reactions (specifically hay fever). The evidence for its use for this purpose indicates its effectiveness but second generation antihistamines such as ketotifen and loratadine have shown equal results with fewer side effects.
It is also used as a preventive treatment against migraine. In a 2013 study the frequency of migraine was dramatically reduced in patients within 7 to 10 days after starting treatment. The average frequency of migraine attacks in these patients before administration was 8.7 times per month, this was decreased to 3.1 times per month at 3 months after the start of treatment. This use is on the label in the UK and some other countries.
It is also used off-label in the treatment of cyclical vomiting syndrome in infants; the only evidence for this use comes from retrospective studies.
Cyproheptadine is sometimes used off-label to improve akathisia in people on antipsychotic medications.
It used off-label to treat various dermatological conditions, including psychogenic itch drug-induced hyperhidrosis (excessive sweating), and prevention of blister formation for some people with epidermolysis bullosa simplex.
One of the effects of the drug is increased appetite and weight gain, which has led to its use (off-label in the USA) for this purpose in children who are wasting as well as people with cystic fibrosis.
It is also used off-label in the management of moderate to severe cases of serotonin syndrome, a complex of symptoms associated with the use of serotonergic drugs, such as selective serotonin reuptake inhibitors (and monoamine oxidase inhibitors), and in cases of high levels of serotonin in the blood resulting from a serotonin-producing carcinoid tumor.
Adverse effects
Adverse effects include:
Sedation and sleepiness (often transient)
Dizziness
Disturbed coordination
Confusion
Restlessness
Excitation
Nervousness
Tremor
Irritability
Insomnia
Paresthesias
Neuritis
Convulsions
Euphoria
Hallucinations
Hysteria
Faintness
Allergic manifestation of rash and edema
Diaphoresis
Urticaria
Photosensitivity
Acute labyrinthitis
Diplopia (seeing double)
Vertigo
Tinnitus
Hypotension (low blood pressure)
Palpitation
Extrasystoles
Anaphylactic shock
Hemolytic anemia
Blood dyscrasias such as leukopenia, agranulocytosis and thrombocytopenia
Cholestasis
Hepatic (liver) side effects such as:
Hepatitis
Jaundice
Liver failure
Hepatic function abnormality
Epigastric distress
Anorexia
Nausea
Vomiting
Diarrhea
Anticholinergic side effects such as:
Blurred vision
Constipation
Xerostomia (dry mouth)
Tachycardia (high heart rate)
Urinary retention
Difficulty passing urine
Nasal congestion
Nasal or throat dryness
Urinary frequency
Early menses
Thickening of bronchial secretions
Tightness of chest and wheezing
Fatigue
Chills
Headache
Increased appetite
Weight gain
Veterinary use
Cyproheptadine is used in cats as an appetite stimulant and as an adjunct in the treatment of asthma. Possible adverse effects include excitement and aggressive behavior. The elimination half-life of cyproheptadine in cats is 12 hours.
Cyproheptadine is a second line treatment for pituitary pars intermedia dysfunction in horses.
————————————————————————–
How to treat COVID, long-haul, and COVID vaccine side-effects (Debated)
Aug 30, 2021
https://www.skirsch.io/how-to-treat-covid/
————————————————————————–
SHOT IN THE ARM: Can I have the Covid vaccine if I’m allergic to penicillin?
Dec 10 2020
https://www.the-sun.com/news/1947844/covid-vaccine-allergic-to-penicillin/
————————————————————————–
Sinus Infections and Antibiotics
May 04, 2021
https://www.verywellhealth.com/antibiotics-for-sinus-infection-5176155
————————————————————————–
Did You Know That Bactrim Can Treat Multiple Infections?
2008
https://www.findatopdoc.com/Healthy-Living/Did-You-Know-That-Bactrim-Can-Treat-Multiple-Infections
Bacteria are microorganisms that thrive in the environment, but they are not always harmful. There are a number of vital functions that are performed by bacteria, which are essential for our day-to-day living. However, sometimes, these bacteria may leave harmful strains on or inside our body. When a bacterium leaves such harmful strains and the person’s immune system cannot fight it off, bacterial infections can take place.
A number of diseases such as food poisoning, meningitis, and pneumonia are a result of such bacterial attacks. The infections caused by these bacteria can be treated using appropriate medications. To fight these bacterial infections, certain antibiotics are prescribed by healthcare providers.
What is Bactrim?
Generic name: Sulfamethoxazole/Trimethoprim
Bactrim is a brand name of an antibiotic that is used to treat many bacterial infections. It is a combination of the two generic antibiotics called sulfamethoxazole and trimethoprim. Both of these antibiotics are used to control various bacterial infections.
Sulfamethoxazole (SMX or SMZ)
Sulfamethoxazole is an effective medication against gram-positive and gram-negative bacteria. This antibiotic can be combined with another antibiotic or as a separate component to treat bacterial infections. Sulfamethoxazole is commonly used as a treatment for urinary and respiratory tract infections, which are bacterial of origin.
Trimethoprim (TMP)
TMP is used for the treatment of urinary bladder infections, traveler’s diarrhea (a stomach and intestinal infection), and middle ear infections.
Uses of Bactrim
Bactrim is used for the treatment of many diseases. They include:
Urinary tract infections (UTI)
Ear infections
Enteritis (inflammation of the small intestine)
Pneumonia
Traveler’s diarrhea
Chronic bronchitis
Pneumonia
Acne
Kidney infection
Prevention of bladder infection
Prostatitis
————————————————————————–
Bactrim
https://www.drugs.com/pro/bactrim.html
————————————————————————–
What Does Bactrim Treat?
https://www.findatopdoc.com/Healthy-Living/what-does-bactrim-treat
————————————————————————–
Sulfa allergy: Which medications should I avoid?
Dec. 04, 2019
https://www.mayoclinic.org/diseases-conditions/drug-allergy/expert-answers/sulfa-allergy/faq-20057970
————————————————————————–
Role of Co-trimoxazole in Severe COVID-19 Patients
2020
https://clinicaltrials.gov/ct2/show/NCT04470531
————————————————————————–
The Role of High Dose Co-trimoxazole in Severe Covid-19 Patients
2021
https://clinicaltrials.gov/ct2/show/NCT04884490
Brief Summary:
Coronavirus Disease 19 (COVID-19) is a worldwide pandemic and a major global health concern which is caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). The newly emerged Coronavirus disease 2019 (COVID-19), which was first identified in Wuhan, China, has swept through 219 countries, killing a staggering number of people. According to WHO reports, the number of deaths had risen to 3,155,168by March 30, 2021, out of 149,910,744 confirmed cases. In Bangladesh, the outbreak has infected over 745,322confirmed cases, with over 11,053 deaths reported. Though the patient may be asymptomatic or present with mild symptoms, mortality is quite high in the severe form of the disease which often progresses to critical phase presented as Acute Respiratory Distress Syndrome (ARDS). This is due to exaggerated response of immune system to the virus termed as cytokine storm syndrome (CSS). There is currently no effective antiviral therapy for SARS-CoV-2 and supportive care is the mainstay of therapy. As a result we are still searching for a better therapeutic agent which will help in treating Covid-19 cases in terms of mortality, morbidity, oxygen requirement, length of stay in hospital. Co-trimoxazole (composed of one-part Trimethoprim and five parts Sulfamethoxazole)is a sulphur containing anti-folate bactericidal drug which is being used for over 60 years for various indications esp. respiratory tract infections. It is known to have immunomodulatory and anti-inflammatory properties that may help to prevent progression to critical phase and cytokine storm syndrome in severe COVID-19 patients. It acts rapidly when given in high dose due to its better bioavailability and lung penetration. Low cost and a good safety profile can make it an ideal candidate for treatment of COVID -19 in a low resource country like Bangladesh.
————————————————————————–
Sulfamethoxazole/trimethoprim: 7 things you should know
Nov 9, 2020
https://www.drugs.com/tips/sulfamethoxazole-trimethoprim-patient-tips
————————————————————————–
Trimethoprim
https://en.wikipedia.org/wiki/Trimethoprim
Trimethoprim (TMP) is an antibiotic used mainly in the treatment of bladder infections. Other uses include for middle ear infections and travelers’ diarrhea. With sulfamethoxazole or dapsone it may be used for Pneumocystis pneumonia in people with HIV/AIDS. It is taken by mouth.
Common side effects include nausea, changes in taste, and rash. Rarely it may result in blood problems such as not enough platelets or white blood cells. Trimethoprim may cause sun sensitivity. There is evidence of potential harm during pregnancy in some animals but not humans. It works by blocking folate metabolism via dihydrofolate reductase in some bacteria which results in their death.
Trimethoprim was first used in 1962. It is on the World Health Organization’s List of Essential Medicines. It is available as a generic medication.
————————————————————————–
TRIMETHOPRIM/SULFAMETHOXAZOLE (TMP/SMX, Bactrim, Septra)
2010
http://www.aids.org/2010/09/trimethoprim-sulfamethoxazole/
WHAT IS TMP/SMX?
TMP/SMX is combination of two antibiotic drugs: trimethoprim and sulfamethoxazole. It is also known as cotrimoxazole. It is sold as Bactrim® (manufactured by Roche) or Septra® (by Monarch Pharmaceuticals). TMP/SMX is sold under many other names in different parts of the world.
Antibiotics fight infections caused by bacteria. TMP/SMX is also used to fight some infections caused by protozoa, and some opportunistic infections in people with HIV.
WHY DO PEOPLE WITH HIV TAKE TMP/SMX?
TMP/SMX is used for many bacterial infections. It is effective and inexpensive. Unfortunately, up to one third of the people who take it get an allergic reaction
Many germs live in our bodies or are common in our surroundings. A healthy immune system can fight them off or keep them under control. However, HIV infection can weaken the immune system. Infections that take advantage of weakened immune defenses are called “opportunistic infections.” People with advanced HIV disease can get opportunistic infections. See Fact Sheet 500 for more information on opportunistic infections.
One opportunistic infection in people with HIV is PCP. This stands for pneumocystis pneumonia, which affects the lungs. See Fact Sheet 515 for more information on PCP. People who have a CD4 cell count of less than 200 may develop PCP.
TMP/SMX is the first choice to treat or prevent PCP. If your CD4 cell count is below 200, ask your health care provider if you should be taking TMP/SMX or another drug to prevent PCP.
Another opportunistic infection is toxoplasmosis (toxo), which affects the brain. See Fact Sheet 517 for more information on toxo. People who have a CD4 cell count of less than 100 may develop toxo. TMP/SMX is sometimes used to treat or prevent cases of toxo.
Some people are allergic to TMP/SMX. Be sure to tell your health care provider if you are allergic to sulfa drugs or antibiotics. People who are anemic should not use TMP/SMX. Taking TMP/SMX during pregnancy may increase the risk of birth defects. Women who are pregnant or breastfeeding should avoid taking it if possible. Also let your health care provider know if you have liver disease, kidney disease, or a shortage of the enzyme glucose-6-phosphate dehydrogenase (G6PD).
————————————————————————–
Bactrim (Sulfamethoxazole and Trimethoprim Oral/Injection)
January 4, 2021
https://www.everydayhealth.com/drugs/bactrim
————————————————————————–
Trimethoprim–sulfamethoxazole and risk of sudden death among patients taking spironolactone
March 03, 2015
https://www.cmaj.ca/content/187/4/E138
————————————————————————–
HIV Patients with Trimethoprim-sulfamethoxazole Intolerance May Tolerate Dapsone
January 5, 2017
https://www.contagionlive.com/view/hiv-patients-with-trimethoprim-sulfamethoxazole-intolerance-may-tolerate-dapsone
————————————————————————–
Avoid Trimethoprim-Sulfamethoxazole for Severe MRSA Infections
2015 July
Clinical question: Is trimethoprim-sulfamethoxazole equivalent to vancomycin for the treatment of severe infections caused by methicillin-resistant Staphyloccus aureus?
Bottom line: Trimethoprim-sulfamethoxazole (TMP-SMX) did not achieve noninferiority as compared with vancomycin for the treatment of severe methicillin-resistant Staphyloccus aureus (MRSA) infections in hospitalized patients, and it may lead to increased mortality in the subset of patients with bacteremia.
https://www.the-hospitalist.org/hospitalist/article/122278/antimicrobial-resistant-infections/avoid-trimethoprim-sulfamethoxazole
————————————————————————–
Sulfamethoxazole and Trimethoprim (Bactrim)
https://www.medicinenet.com/sulfamethoxazole_and_trimethoprim/article.htm#what_is_sulfamethoxazole_and_trimethoprim_and_how_does_it_work_mechanism_of_action
————————————————————————–
What Conditions does SULFAMETHOXAZOLE-TRIMETHOPRIM Treat?
Infection due to the bacteria Vibrio cholerae
Typhoid fever
Paratyphoid fever
Intestine infection due to the Shigella bacteria
Prevention of plague following exposure to plague
Infection due to a Brucella bacteria
Brain/spinal cord infection due to Listeria monocytogenes
Whooping cough
Colonization with Meningococcus bacteria without symptoms of infection
Infection due to Nocardia bacteria
Whipple’s disease
Infection due to Chlamydiae species bacteria
Urinary tract infection prevention
Lymphogranuloma venereum
Granuloma inguinale
Paracoccidioidomycosis
Prevention of toxoplasmosis
AIDS with toxoplasmosis
Pneumonia with a fungus called Pneumocystis jirovecii
Pneumocystis jiroveci pneumonia prevention
An infection
Infection of the middle ear by H. influenzae bacteria
Infection of the middle ear caused by Streptococcus
A bacterial infection of the middle ear
Bacterial infection of heart valve
Inflammation of the tissue lining the sinuses
Pneumonia caused by bacteria
Bacterial infection with chronic bronchitis
Chronic bronchitis caused by Streptococcus pneumoniae
Chronic bronchitis caused by Haemophilus influenzae
Diverticulitis
Bacterial urinary tract infection
Infection of urinary tract due to Enterobacter cloacae
Urinary tract infection due to E. coli bacteria
Urinary tract infection caused by Klebsiella bacteria
Infection of the urinary tract from Proteus bacteria
Urinary tract infection caused by Morganella morganii
Continuous bacterial inflammation of the prostate gland
An infection of the skin and the tissue below the skin
Diabetic foot infection
Infection of bone
Infection due to the microorganism Coxiella burnetii
Short-term infection with diarrhea, discomfort & weight loss
https://www.webmd.com/drugs/2/drug-3067-9071/sulfamethoxazole-tmp-ds-oral/sulfamethoxazole-trimethoprim-oral/details/list-conditions
————————————————————————–
Trimethoprim use for urinary tract infection and risk of adverse outcomes in older patients: cohort study
09 February 2018
https://www.bmj.com/content/360/bmj.k341
————————————————————————–
What to know about sulfa allergies
March 28, 2018
https://www.medicalnewstoday.com/articles/321349
————————————————————————–
Renal Transplant Recipient with Concurrent COVID-19 and Stenotrophomonas maltophilia Pneumonia Treated with Trimethoprim/Sulfamethoxazole Leading to Acute Kidney Injury: A Therapeutic Dilemma
2020
https://pubmed.ncbi.nlm.nih.gov/32799217/
————————————————————————–
Metronidazole; a Potential Novel Addition to the COVID-19 Treatment Regimen
2020
https://pubmed.ncbi.nlm.nih.gov/32259129/
————————————————————————–
This new antibody can stop all COVID-19 strains, including new variants, experts say
Aug 27, 2021
https://www.deseret.com/coronavirus/2021/8/27/22643254/antibody-stops-covid-19-coronavirus-variants-delta-lambda
A new antibody theory can reportedly neutralize all COVID-19 strains and coronavirus, paving the way for stopping COVID-19
A team of researchers may have found an antibody that can neutralize all known novel coronavirus strains, including the developing variants.
GlaxoSmithKline and Vir Biotechnology recently conducted a huge collaborative study by scientists and developed a new antibody therapy, called Sotrovimab. During the project, they discovered a new natural antibody “that has remarkable breadth and efficacy,” according to the Berkeley Lab.
The scientists reportedly discovered a new antibody, called S309, which “neutralizes all known SARS-CoV-2 strains — including newly emerged mutants that can now ‘escape’ from previous antibody therapies — as well as the closely related original SARS-CoV virus,” according to a press release from the Berkeley Lab.
Structural biologist Jay Nix, who was involved with the project, said the antibody can potentially stop all coronaviruses similar to COVID-19.
————————————————————————–
All 37 Omicron Mutations Defeated By This New Antibody Treatment
2021
Tests showed that sotrovimab, the drugmaker’s antibody treatment, stands up to all mutations in the spike protein of the omicron variant and not just the key mutations, GlaxoSmithKline Plc said.
https://www.ndtv.com/world-news/all-37-covid-19-omicron-mutations-defeated-by-this-new-antibody-treatment-2641943
————————————————————————–
There could be one COVID-19 vaccine to stop all the variants, experts say
Aug 24, 2021
https://www.deseret.com/coronavirus/2021/8/24/22639561/covid-vaccine-to-stop-coronavirus-variants
Researchers are seeking out one COVID-19 vaccine to stop COVID-19 variants
Could there be one COVID-19 vaccine to rule them all? A number of researchers are trying to discover a COVID-19 vaccine that will stop any and all coronavirus variants that pop up in the future.
Will there be one COVID-19 vaccine to stop variants?
Per Reuters, there are two separate research teams that are conducting tests to create a new vaccine that could stop variants.
One study from the New England Journal of Medicine suggested that there were “high-level, broad-spectrum” antibodies in blood samples from those who survived the SARS outbreak in 2003.
These antibodies could allegedly stop COVID-19 and “also five viruses that have been identified in bats and pangolins and that have the potential to cause human infection,” according to Reuters.
A second study, which was published in the medical journal Immunity, found an antibody that could protect against a number of COVID-19 variants in mice.
“The antibody attaches to a part of the virus that differs little across the variants, meaning that it is unlikely for resistance to arise at this spot,” the authors said, according to Reuters.
Will there be an ideal COVID-19 vaccine?
Back in April, scientists told NPR they’re interested in creating an ideal COVID-19 vaccine that would be given out in a single shot and wouldn’t cause too many problems.
The ideal COVID-19 vaccine would be “administered in a single shot, be room temperature stable, work in all demographics and, even pushed beyond that, ideally be self-administered,” Deborah Fuller, a vaccine researcher at the University of Washington, told NPR.
————————————————————————–
Fauci Was Duplicitous on the AIDS Epidemic Too
February 23, 2021
https://www.aier.org/article/fauci-was-duplicitous-on-the-aids-epidemic-too/
————————————————————————–
Kim Iversen: Is Fauci’s Botched Handling Of The AIDS Epidemic Being Repeated?
Sep 7, 2021
https://www.youtube.com/watch?v=ezKb_AFvU4g
————————————————————————–
Whitewashing AIDS History
02/21/2014
https://www.huffpost.com/entry/whitewashing-aids-history_b_4762295
Dr. Anthony Fauci is rewriting history. He is doing so to disguise his shameful role in delaying promotion of an AIDS treatment that would have prevented tens of thousands of deaths in the first years of the epidemic.
In my book, Body Counts, A Memoir of Politics, Sex, AIDS, and Survival, I recount how slow the federal government was in publicizing the use of Bactrim and other sulfa drugs to prevent PCP (the pneumonia that was then the leading killer of people with AIDS) in addition to its long-time and well-known use to treat PCP.
I point to Dr. Fauci in particular, because he was, and remains today, the head of the National Institutes of Allergy and Infectious Diseases and the head of the federal government’s AIDS research program. In 1987, pioneering AIDS activist Michael Callen begged Fauci for help in promoting the use of Bactrim as PCP prophylaxis and issuing interim guidelines urging physicians to prophylax those patients deemed at high risk for PCP.
Fauci has taken exception to my account of the incident. In comments to The Washington Blade, he offered his version of the story.
“So what actually happened is that Michael came to me and said you know there is this preliminary activity and some small trials that Bactrim works,” Fauci said. “Would you come out and make a guideline to say it should be used by everybody. And I said ‘Michael I can’t do that but what I can do is help design and make sure that the grantees that we fund do a clinical trial in Bactrim to prove or not that it was safe and effective,’” he said. “But I didn’t blow him off and say I don’t want to issue guidelines. The fact is that’s neither within my purview nor within the responsibility or authority I have to issue guidelines.”
Whether Fauci personally had this authority or not isn’t the point. His excuse would be laughable if it wasn’t so tragic; he was constantly traveling and speaking to the media and opining about everything related to AIDS research and treatment. In Arthur Kahn’s book, Winter Wars, Larry Kramer pointed out that to get an appointment with Fauci, one didn’t call his secretary but his press officers, “who book [his] talks and interviews… like movie stars.” He could easily have advocated awareness of the preventive treatment, as the de facto federal AIDS Czar, his influence was and is enormous.
Had Fauci listened to people with AIDS and the clinicians treating them, and responded accordingly, he would have saved thousands of lives. In the two years between 1987, when Callen met with Fauci and 1989, when the guidelines were ultimately issued, nearly 17,000 people with AIDS suffocated from PCP. Most of these people might have lived had Fauci responded appropriately.
When Callen and others, including Dr. Barry Gingell, a medical advisor to Gay Men’s Health Crisis, met with Fauci to plead for his support, they didn’t just say there was “this preliminary activity and some small trials,” as Fauci claims. They explained that many frontline AIDS physicians, following the lead of Dr. Joseph Sonnabend, were already using Bactrim effectively to prevent the recurrence of PCP. The science was clear. A decade before, clinical trials by Dr. Walter Hughes had proven its efficacy in preventing PCP in other immune-compromised populations, like children with leukemia.
Hughes suggested that Bactrim prophylaxis should be used whenever the recurrence rate of PCP was over 15 percent. Sonnabend and community clinicians had demonstrated that the recurrence rate of PCP in AIDS was over 60 percent in the year following the first episode. If ever there was a group in urgent need of a recommendation for Bactrim prophylaxis, it was people with AIDS.
Fauci refused to acknowledge the evidence and, according to one account, even encouraged people with AIDS to stop taking treatments, like Bactrim, that weren’t specifically approved for use in people with AIDS. Longtime treatment activist Richard Jefferys wrote in 2001 that Fauci “went as far as telling activists attending a 1987 meeting that there was no data to suggest PCP prophylaxis was beneficial and that it may, in fact be dangerous.” Fauci’s close colleague, Dr. Samuel Broder, who was head of the National Cancer Institute, even suggested — in the absence of any evidence at all — that the newly introduced antiretroviral, AZT, would make prophylaxis against PCP redundant!
In his comment to the Blade, Fauci fails to mention how skeptical he was of Bactrim as a preventive treatment, that he questioned the existing science, was unswayed by how frontline clinicians were treating people with AIDS and had suggested stopping a treatment that was already saving lives.
Sonnabend wrote in 2006, “Why, in the case of AIDS, was Bactrim, a known preventative measure against PCP, introduced so many years after a need for it had been recognized? To this must be added the question of why this neglect, the consequences of which can be measured in the tens of thousands of lives lost, has received almost no attention.”
If we don’t tell the truth about the history of the AIDS epidemic, it will be subject to more whitewashing, more distortions and more rewriting to suit the legacies of the officials in charge.
These are the same officials who seem incapable of ever acknowledging or taking responsibility for mistakes they made — mistakes that cost our community thousands of lives.
————————————————————————–
Through AIDS, Ebola and Covid, Dr. Fauci Is Still Treating Patients
Dec 24, 2020
https://www.wsj.com/articles/anthony-fauci-doctor-covid-vaccine-ebola-11608776073
————————————————————————–
Could gene therapies be used to cure more people with HIV?
31 August 2021
https://www.aidsmap.com/news/aug-2021/could-gene-therapies-be-used-cure-more-people-hiv
————————————————————————–
Study paves the way for development of novel delivery systems to suppress HIV
Sep 21 2021
In a new study supported by the National Institutes of Health, researchers used exosomes, tiny nanoparticles capable of being taken up by cells, to deliver novel protein into the cells of mice infected with HIV. The protein attached to HIVs’ genetic material and prevented it from replicating, resulting in reduced levels of HIV in the bone marrow, spleen, and brain. The study, funded by NIH’s National Institute of Mental Health (NIMH) and published in Nature Communications, paves the way for the development of novel delivery systems for suppressing HIV.
“These results demonstrate the potential of exosome engineering for delivering epigenetics-based therapeutics capable of silencing HIV gene expression into brain tissues – an area where HIV has traditionally been able to hide from HIV treatments.”
Jeymohan Joseph, Ph.D., Chief, HIV Neuropathogenesis, Genetics, and Therapeutics Branch, NIMH’s Division of AIDS Research
HIV attacks the immune system by infecting a type of white blood cell in the body that is vital to fighting off infection. Without treatment, HIV can destroy these white blood cells, reducing the body’s ability to mount an immune response—eventually resulting in AIDS. Although researchers have been working to develop new therapies to treat and cure HIV and AIDS, this quest is challenging for many reasons. One reason is that HIV can enter a dormant-like state, hiding in the body and evading treatments, only to reactivate at a later date. HIV hiding in the brain is particularly difficult to access, as the blood-brain barrier often prevents treatments from entering into those tissues.
One avenue researchers have been pursuing in their efforts to try to cure HIV is what is sometimes called a “block and lock” approach, particularly for targeting HIV in the brain. This method attempts to block the virus’ ability to replicate itself and lock it in its dormant state.
Kevin Morris, Ph.D., of City of Hope and the Menzies Health Institute Queensland at Griffith University, Australia, led an investigation into a new approach for blocking and locking HIV in mice. The researchers use exosomes, tiny nanoparticles capable of being taken up by cells, to deliver a novel recombinant anti-HIV protein, called ZPAMt, into cells infected with HIV. The ZPAMt protein was designed by researchers to attach to a region of the virus called LTR that is critical for virus replication. The protein has an epigenetic marker in it that changes the way HIVs’ genetic information is expressed, suppressing it, and making the virus unable to divide and multiply. The exosomes are able to cross the blood brain barrier and enter into the brain making this treatment capable of targeting this hard-to-reach organ.
https://www.news-medical.net/news/20210921/Study-paves-the-way-for-development-of-novel-delivery-systems-to-suppress-HIV.aspx
————————————————————————–
A common cold virus may help fight COVID-19
March 29, 2021
https://www.medicalnewstoday.com/articles/a-common-cold-virus-may-help-fight-covid-19
A lab-based study has found that a virus that causes the common cold can trigger an innate immune response against SARS-CoV-2, the virus responsible for COVID-19.
In theory, infections with the common cold virus could inhibit the transmission of SARS-CoV-2 among members of a population and reduce the severity of infections.
Further research could lead to control strategies or treatments that exploit such interactions between viruses.
————————————————————————–
If You’ve Had This Illness, You May Be Immune to COVID-19
6/11/2020
https://www.msn.com/en-us/health/medical/if-you-ve-had-this-illness-you-may-be-immune-to-covid-19/ar-BB15mj7i
Gov. Kate Brown expected to move 14 counties to ‘extreme risk,’ shutter…
Miami School Says It Won’t Employ Vaccinated Teachers, Citing…
Decades before the terms “COVID-19” or “novel coronavirus” started dominating headlines around the world, there was SARS, aka severe acute respiratory syndrome, an illness caused by coronavirus. The condition was first reported in China in 2002 and spread around the world within a few months. Luckily, it was contained pretty quickly, and no known transmissions have occurred since 2004. However, if you are one of the 8,098 people worldwide who became infected with the virus, you may have immunity against coronavirus, according to a new study published in the medical journal Nature.
May Have Antibodies That Fight Coronavirus
In the paper, researchers San Francisco’s Vir Biotechnology and the University of Washington, explain that when examining old blood samples from an individual who had been infected with SARS coronavirus in 2003, they discovered an antibody—S309—in the blood of a person that effectively blocked SARS-CoV-2. When they attempted to isolate the antibody and then add the virus, SARS-CoV-2 wasn’t able to enter cells and replicate. While scientists assumed that the antibodies would have some commonalities, since the two viruses are closely related, they were surprised to find how potent the SARS antibodies actually were. The team is still trying to figure out why S309 effectively blocks the virus.
“Looking for effective antibodies is like looking for a needle in a haystack,” David Veesler, a senior author on the paper and a virologist at the University of Washington, told the San Francisco Chronicle. “So this was very, very exciting because this antibody has the potential to have a high public health impact.”
————————————————————————–
Could Covid-19 immunity last for 17 YEARS? Researchers find SARS patients still have crucial T cells from when they were infected in 2003
July 2020
The SARS epidemic started in 2003. No cases have been identified since 2004
Some infected people during the outbreak still have crucial white T cells
It suggests they would be protected from SARS re-infection
It offers hope the same could be true for this virus, called SARS-2
https://www.dailymail.co.uk/news/article-8529429/Could-immunity-17-YEARS-Singaporean-researchers-SARS-patients-crucial-T-cells.html
————————————————————————–
Adaptive immunity to SARS-CoV-2 and COVID-19
2021
https://pubmed.ncbi.nlm.nih.gov/33497610/
Abstract
The adaptive immune system is important for control of most viral infections. The three fundamental components of the adaptive immune system are B cells (the source of antibodies), CD4+ T cells, and CD8+ T cells. The armamentarium of B cells, CD4+ T cells, and CD8+ T cells has differing roles in different viral infections and in vaccines, and thus it is critical to directly study adaptive immunity to SARS-CoV-2 to understand COVID-19. Knowledge is now available on relationships between antigen-specific immune responses and SARS-CoV-2 infection. Although more studies are needed, a picture has begun to emerge that reveals that CD4+ T cells, CD8+ T cells, and neutralizing antibodies all contribute to control of SARS-CoV-2 in both non-hospitalized and hospitalized cases of COVID-19. The specific functions and kinetics of these adaptive immune responses are discussed, as well as their interplay with innate immunity and implications for COVID-19 vaccines and immune memory against re-infection.
————————————————————————–
What Scientists Know About Immunity to the Novel Coronavirus
March 30, 2020
Though COVID-19 likely makes recovered patients immune, experts aren’t sure how long protection lasts
https://www.smithsonianmag.com/science-nature/can-you-become-immune-sars-cov-2-180974532/
————————————————————————–
Hyperimmune response model could clarify severe COVID-19
January 22nd, 2021
https://www.futurity.org/hyperimmune-response-sars-cov-2-covid-19-2505382-2/
————————————————————————–
Scientists uncover SARS-CoV-2-specific T cell immunity in recovered COVID-19 and SARS patients
July 16, 2020
https://medicalxpress.com/news/2020-07-scientists-uncover-sars-cov-specific-cell-immunity.html
————————————————————————–
Functional impairment of HIV-specific T cells precedes aborted virus control
Sep 13 2021
https://www.news-medical.net/news/20210913/Functional-impairment-of-HIV-specific-T-cells-precedes-aborted-virus-control.aspx
————————————————————————–
Memory killer T cells are primed in the spleen during influenza infection
September 10, 2021
https://medicalxpress.com/news/2021-09-memory-killer-cells-primed-spleen.html
————————————————————————–
Prior SARS-CoV-2 infection rescues B and T cell responses to variants after first vaccine dose
25 Jun 2021
https://science.sciencemag.org/content/372/6549/1418.full
————————————————————————–
T cells can counteract dangerous phenomenon of mosquito-borne viruses
Jun 5 2020
https://www.news-medical.net/news/20200605/T-cells-can-counteract-dangerous-phenomenon-of-mosquito-borne-viruses.aspx
————————————————————————–
New research: Mosquito protein inhibits number of viruses, raises hope against Covid too
March 16, 2021
A mosquito protein, called AEG12, strongly inhibits the family of viruses that cause yellow fever, dengue, West Nile, and Zika, and also weakly inhibits coronaviruses, according to new research.
https://indianexpress.com/article/explained/mosquito-protein-inhibits-number-of-viruses-raises-hope-against-covid-too-7228366/
————————————————————————–
Presentation of fatal stroke due to SARS-CoV-2 and dengue virus coinfection
2020 Dec 30
https://pubmed.ncbi.nlm.nih.gov/32881018/
————————————————————————–
COVID-19’s impact on dengue transmission
November 4, 2020
https://www.medicalnewstoday.com/articles/covid-19s-impact-on-dengue-transmission
————————————————————————–
The role of natural killer cells in fight against viral infections including SARS-CoV-2
Jun 17 2021
Natural killer (NK) cells are a subpopulation of lymphocytes that have various roles within the immune system. To this end, NK cells have unique cell recognition capabilities and have been shown to play a role in controlling viral and intracellular bacterial infections and tumors.
Today, NK cells are used in various therapies, including those to treat cancer and certain viral diseases. Recent studies have found a role of NK cells in fighting coronavirus disease (COVID-19), which is caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
In an effort to further understand the role of NK cells in combatting COVID-19, researchers at the Karolinska University Hospital in Sweden explored NK cells in acute viral infections, citing flavivirus and influenza virus infections as examples. The researchers also studied the activation of NK cells and NK cell function in humans infected with SARS-CoV-2.
Their study, which was published in Nature Reviews Immunology, highlights the role of NK cells in acute viral infections. This data can shed light on how NK cells emerge when a person has COVID-19, which has now infected over 177 million people worldwide.
Natural killer (NK) cells
NK cells are widely known for their cytotoxic function and ability to induce the production of cytokines when stimulated. This population of lymphocytes typically targets infected, transformed, and stressed cells, thereby allowing NK cells to eliminate unwanted cells while also contributing to cellular homeostasis.
NK cells primarily originate from the bone marrow, which is where these cells interact with other cellular components and cytokines to support their development. Recent studies have shown that in addition to the bone marrow, NK cells can also arise from the peripheral blood and secondary lymphoid organs, as well as specific tissues called tissue-resident NK cells. In humans, NK cells are identified as CD3-CD56+ lymphocytes.
During a viral infection, the host cells may become vulnerable to NK cell-mediated recognition through the upregulation of self-encoded molecules triggered by the infection or a cellular stress response. These molecules will then bind to activating NK cell receptors like natural cytotoxicity receptors, C-type lectin-like receptors, and co-activating receptors. Another role of NK cells is to remove virus-infected cells through the CD16-mediated antibody-dependent cellular cytotoxicity (ADCC).
Taken together, NK cell activity is largely modulated by cytokines, including the interleukin-2 (IL-2), IL-12, IL-15, IL-18, and type I interferons. These cytokines, either alone or in combination, can contribute to NK cell survival, proliferation, and cytotoxicity, as well as the ability of NK cells to produce cytokines like interferon-γ (IFN-γ).
NK cell responses to flaviviruses
In their Nature Reviews Immunology study, the investigators reviewed current knowledge about human NK cells in antiviral immunity. During acute infections with flavivirus and influenza virus, which are both RNA viruses, NK cells respond strongly. Viral infections stimulate the activation of circulating NK cells, thus attracting these lymphocytes to the affected tissues. Tissue-resident NK cells can also become activated upon the recognition of these infections, which also plays a role in supporting the immune system’s response.
Flaviviruses such as the dengue virus (DENV), West Nile virus, tick-borne encephalitis virus, Japanese encephalitis virus, Zika virus, and the Yellow fever virus are major emerging hemorrhagic-like syndromes and central nervous system-linked infections. Though there are several vaccines that are currently available to prevent some of these infections, there remains a lack of knowledge regarding which host cells are affected by flaviviruses in humans and whether they are targets of NK cell-mediated responses.
————————————————————————–
Too much of a good thing? Overactive immune response blocks itself
July 26, 2013
As part of the innate immune system natural killer cells (NK cells) play an important role in immune responses. For a long time they have been known as the first line of defense in the fight against infectious diseases. Therefore, researchers assumed that the body needs as many active NK cells as possible. However, scientists at the Helmholtz Centre for Infection Research (HZI) have now shown that the principle “the more the better” does not apply to this type of immune cells.
https://www.sciencedaily.com/releases/2013/07/130726103355.htm
————————————————————————–
SARS-CoV-2 RNA-dependent RNA polymerase as a therapeutic target for COVID-19
2021 Mar 3
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7938656/
————————————————————————–
TMEM41B Is a Pan-flavivirus Host Factor
2020
Abstract
Flaviviruses pose a constant threat to human health. These RNA viruses are transmitted by the bite of infected mosquitoes and ticks and regularly cause outbreaks. To identify host factors required for flavivirus infection, we performed full-genome loss of function CRISPR-Cas9 screens. Based on these results, we focused our efforts on characterizing the roles that TMEM41B and VMP1 play in the virus replication cycle. Our mechanistic studies on TMEM41B revealed that all members of the Flaviviridae family that we tested require TMEM41B. We tested 12 additional virus families and found that SARS-CoV-2 of the Coronaviridae also required TMEM41B for infection. Remarkably, single nucleotide polymorphisms present at nearly 20% in East Asian populations reduce flavivirus infection. Based on our mechanistic studies, we propose that TMEM41B is recruited to flavivirus RNA replication complexes to facilitate membrane curvature, which creates a protected environment for viral genome replication.
https://pubmed.ncbi.nlm.nih.gov/33338421/
————————————————————————–
Flavivirus infection requires specific host genes
June 28, 2016
Flaviviruses—a family that includes the Zika, West Nile, Japanese encephalitis, dengue, and yellow fever viruses—are typically transmitted by mosquitoes or ticks. These viruses infect hundreds of millions of people each year. They cause a wide range of symptoms, such as fever, shock, meningitis, paralysis, birth defects, and death.
Since viruses rely on their infected host to create viral particles that spread infection, a research team led by Dr. Michael S. Diamond at Washington University in Saint Louis, Missouri carried out a genome-wide screen to identify host genes necessary for flavivirus infections. Using the CRISPR/Cas9 genetic editing system, they individually removed more than 19,000 genes from human cells and screened for cells that survived West Nile Virus infection. The work was funded in part by NIH’s National Institute of Allergy and Infectious Diseases (NIAID). Results were published online in Nature on June 17, 2016.
The researchers identified 9 genes that, when removed, reduced West Nile infection in the cells. All are associated with the cell’s endoplasmic reticulum, where viral translation, replication, and assembly of viral particles takes place. Six of the genes also reduced infection of Zika, dengue, Japanese encephalitis, and yellow fever viruses when removed from human cells. Most showed similar effects when absent from insect cells tested for West Nile and dengue virus infections.
Two of the identified genes are components of the signal peptidase complex (SPC). The SPC processes proteins for entry into the endoplasmic reticulum. Genetically removing the Signal Peptidase Complex Subunit 1 (SPCS1) gene in human cells reduced levels of all flaviviruses tested (West Nile, dengue, Zika, yellow fever, Japanese encephalitis, and hepatitis C viruses), but had little effect on other viruses (alphavirus, bunyavirus, or rhabdovirus).
Three flavivirus proteins (C, prM, and E) are needed during the viral particle assembly process. These are cleaved from a single flavivirus precursor protein upon entering the endoplasmic reticulum. In cells lacking SPCS1, levels of prM and E were reduced and correlated with a lower number of viral particles. This suggests that SPCS1 may regulate protein processing of flaviviruses.
“Flaviviruses appear to be uniquely dependent on this particular gene to assemble their viral particles,” Diamond says. “In these viruses, this gene sets off a domino effect that is required to assemble and release the viral particle. Without it, the chain reaction doesn’t happen, and the virus can’t spread.”
The findings suggest that SPCS1 could be a potential therapeutic target for stopping flavivirus infections. Notably, an accompanying study in Nature used a similar approach to screen for host factors needed by the dengue and Hepatitis C viruses. That study uncovered some of the same, as well as other, host factors necessary for viral replication. Together, these studies suggest several potential new targets for antiviral therapies.
https://www.nih.gov/news-events/nih-research-matters/flavivirus-infection-requires-specific-host-genes
————————————————————————–
The continued threat of emerging flaviviruses
04 May 2020
https://www.nature.com/articles/s41564-020-0714-0
————————————————————————–
Covid-19: Molnupiravir reduces risk of hospital admission or death by 50% in patients at risk, MSD reports
09 October 2021
https://www.bmj.com/content/375/bmj.n2422/rr-5
————————————————————————–
Buyer beware: molnupiravir may damage DNA
04 November 2021
Covid-19: Molnupiravir effects
https://www.bmj.com/content/375/bmj.n2663
————————————————————————–
ICMR’s thumbs down to Covid drug Molnupiravir leaves medical fraternity divided
January 2021
https://www.msn.com/en-in/health/medical/icmr-s-thumbs-down-to-covid-drug-molnupiravir-leaves-medical-fraternity-divided/ar-AASMtYn
————————————————————————–
Molnupiravir
https://en.wikipedia.org/wiki/Molnupiravir
Molnupiravir is an antiviral medication that inhibits the replication of certain RNA viruses. It is used to treat COVID-19 in those infected by SARS-CoV-2.
Molnupiravir is a prodrug of the synthetic nucleoside derivative N4-hydroxycytidine and exerts its antiviral action through introduction of copying errors during viral RNA replication.
Molnupiravir was originally developed to treat influenza at Emory University by the university’s drug innovation company, Drug Innovation Ventures at Emory (DRIVE), but was reportedly abandoned for mutagenicity concerns. It was then acquired by Miami-based company Ridgeback Biotherapeutics, which later partnered with Merck & Co. to develop the drug further.
Based on positive results in placebo-controlled double-blind randomized clinical trials, Molnupiravir was approved for medical use in the United Kingdom in November 2021. In December 2021, the U.S. Food and Drug Administration (FDA) granted an emergency use authorization (EUA) to molnupiravir for use in certain populations where other treatments are not feasible. The emergency use authorization was only narrowly approved (13-10) because of questions regarding efficacy and drug safety. A prominent virologist made unsubstantiated claims that molnupiravir’s mutagenic effects could create new variants that evade immunity and prolong the COVID-19 pandemic.
Medical uses
Molnupiravir is indicated for the treatment of mild-to-moderate coronavirus disease (COVID-19) in adults with positive results of direct SARS-CoV-2 viral testing, and who are at high risk for progression to severe COVID-19.
————————————————————————–
Thailand and Indonesia unveil plans to develop molnupiravir COVID-19 pill
January 14, 2022
https://www.reuters.com/world/asia-pacific/thailand-indonesia-unveil-plans-develop-molnupiravir-covid-19-pill-2022-01-14/
————————————————————————–
Potential DNA vaccine integration into host cell genome
1995 Nov
https://pubmed.ncbi.nlm.nih.gov/8546411/
————————————————————————–
Cell-Mediated Immune Responses to COVID-19 Infection
July 2020
https://www.frontiersin.org/articles/10.3389/fimmu.2020.01662/full
————————————————————————–
It Turns Out, We All May Have Some Immunity to COVID, New Study Shows
Recent research proves that our cells can help protect us from coronavirus and other pathogens.
July 23, 2020
https://bestlifeonline.com/coronavirus-immunity/
————————————————————————–
Coronavirus
https://en.wikipedia.org/wiki/Coronavirus
(Transmission electron micrograph (TEM) of avian infectious bronchitis virus).
(Illustration of the morphology of coronaviruses; the club-shaped viral spike peplomers, colored red, create the look of a corona surrounding the virion when observed with an electron microscope).

(Cross-sectional model of a coronavirus).
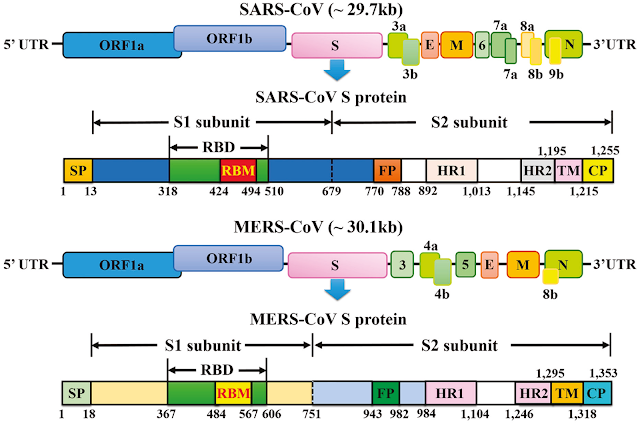
(Schematic representation of the genome organization and functional domains of S protein for SARS-CoV and MERS-CoV).

(The life cycle of a coronavirus).
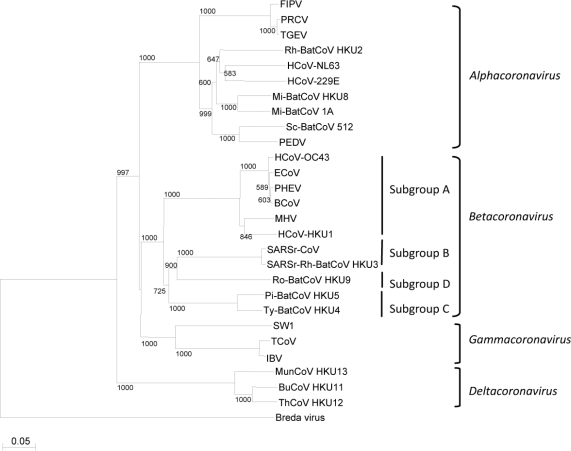
(Phylogenetic tree of coronaviruses.)
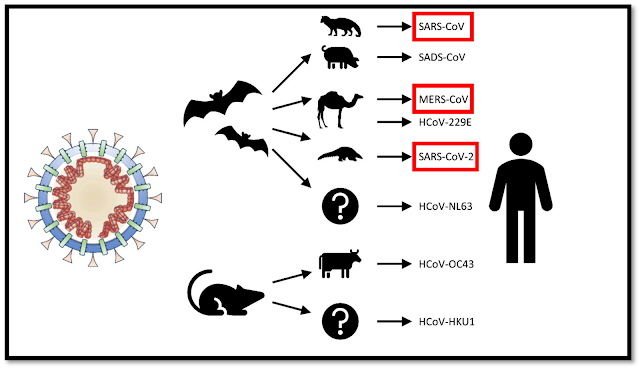
(Origins of human coronaviruses with possible intermediate hosts.)
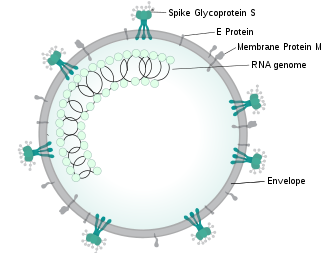
(Illustration of SARSr-CoV virion).
| MERS-CoV | SARS-CoV | SARS-CoV-2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Disease | MERS | SARS | COVID-19 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Outbreaks | 2012, 2015, 2018 | 2002–2004 | 2019–2020 pandemic | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Epidemiology | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Date of first identified case | June 2012 | November 2002 | December 2019 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Location of first identified case | Jeddah, Saudi Arabia | Shunde, China | Wuhan, China | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Age average | 56 | 44[55][a] | 56 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sex ratio (M:F) | 3.3:1 | 0.8:1[57] | 1.6:1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Confirmed cases | 2494 | 8096[58] | 1,981,239 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Deaths | 858 | 774[58] | 126,681 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Case fatality rate | 37% | 9.2% | 6.4% | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symptoms | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Fever | 98% | 99–100% | 87.9% | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Dry cough | 47% | 29–75% | 67.7% | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Dyspnea | 72% | 40–42% | 18.6% | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Diarrhea | 26% | 20–25% | 3.7% | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sore throat | 21% | 13–25% | 13.9% | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Ventilatory use | 24.5%[61] | 14–20% | 4.1% | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
————————————————————————
————————————————————————
————————————————————————
How China’s “Bat Woman” Hunted Down Viruses from SARS to the New Coronavirus
March 11, 2020
Wuhan-based virologist Shi Zhengli has identified dozens of deadly SARS-like viruses in bat caves, and she warns there are more out there
https://www.scientificamerican.com/article/how-chinas-bat-woman-hunted-down-viruses-from-sars-to-the-new-coronavirus1/
—————————–
Uh Oh…The Wuhan Lab-Leak Debate Just Got EVEN MESSIER!!!
Oct 3, 2021
https://www.youtube.com/watch?v=yTbzyirwnoQ
—————————–
Ecological dynamics of emerging bat virus spillover
Abstract
Viruses that originate in bats may be the most notorious emerging zoonoses that spill over from wildlife into domestic animals and humans. Understanding how these infections filter through ecological systems to cause disease in humans is of profound importance to public health. Transmission of viruses from bats to humans requires a hierarchy of enabling conditions that connect the distribution of reservoir hosts, viral infection within these hosts, and exposure and susceptibility of recipient hosts. For many emerging bat viruses, spillover also requires viral shedding from bats, and survival of the virus in the environment. Focusing on Hendra virus, but also addressing Nipah virus, Ebola virus, Marburg virus and coronaviruses, we delineate this cross-species spillover dynamic from the within-host processes that drive virus excretion to land-use changes that increase interaction among species. We describe how land-use changes may affect co-occurrence and contact between bats and recipient hosts. Two hypotheses may explain temporal and spatial pulses of virus shedding in bat populations: episodic shedding from persistently infected bats or transient epidemics that occur as virus is transmitted among bat populations. Management of livestock also may affect the probability of exposure and disease. Interventions to decrease the probability of virus spillover can be implemented at multiple levels from targeting the reservoir host to managing recipient host exposure and susceptibility.
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4262174/
——————————
‘Could have been much worse’: India stops deadly Nipah outbreak
September 2018
https://www.healio.com/infectious-disease/zoonotic-infections/news/print/infectious-disease-news/%7Bb8d4593f-f642-430d-aecf-867ca00df25e%7D/could-have-been-much-worse-india-stops-deadly-nipah-outbreak
Experts credited a strong public health response with containing a deadly outbreak of Nipah virus in India in the same month that it began.
Nineteen patients were infected in the outbreak in the state of Kerala — most in the hospital — and 17 died, including at least one health care worker, officials said. Soon after the outbreak was reported to the National Center for Disease Control on May 19, India launched a coordinated response to contain the spread that included local, state and national health agencies. No new cases were reported after May 29.
——————————
What Is Nipah Virus? Outbreak in India Kills at Least 10
May 21, 2018
The Nipah virus in a sample of cerebrospinal fluid from an infected patient.
https://www.livescience.com/62627-nipah-virus-outbreak-india.html
——————————-
Nipah Outbreak In India Leaves 17 Dead
Since the outbreak first started on May 19, 2018, there have been 17 deaths from Nipah virus infections in Kerala, India.
https://www.asianscientist.com/2018/06/health/kerala-india-nipah-virus-outbreak/
——————————-
How India Stopped a Deadly Brain-Damaging Virus Caused by Flying Fox Drool in 2018
3-14-2020
You thought Coronavirus was scary?
The deadly virus first reared its pointy-eared head in 1998/1999 when pig farmers in Nipah, Malaysia began succumbing to fevers accompanied by flu-like symptoms. Survivors sometimes exhibited altered personalities due to brain damage.
In fact, Tthe Nipah virus is carried by (but does not affect) fruit bats, who can transmit the disease in their saliva or urine. The pigs contacted fruit bat saliva from fruit fallen from trees infested by the bats, and the farmers were then infected due to their contact with the fluids from the pigs. Ultimately a million pigs were slaughtered in Malaysia to contain the disease, which infected at least 265 persons and killed 105.
https://nationalinterest.org/blog/buzz/how-india-stopped-deadly-brain-damaging-virus-caused-flying-fox-drool-2018-133267
——————————-
Bat-borne Nipah virus could help explain COVID-19
December 4, 2020
https://scopeblog.stanford.edu/2020/12/04/bat-borne-nipah-virus-could-help-explain-covid-19/
——————————-
Brain-swelling Nipah virus 75 times more deadly than coronavirus may be next pandemic, scientists warn of ‘The Big One’
February 21, 2021
A brain-swelling disease 75 times more deadly than coronavirus could become the next pandemic killing millions, scientists have warned.
https://www.news.com.au/lifestyle/health/health-problems/brainswelling-nipah-virus-75-times-more-deadly-than-coronavirus-may-be-next-pandemic-scientists-warn-of-the-big-one/news-story/5363e34783bd54c926434c738edd5e12
——————————-
Post-COVID: Let’s Prevent the Nipah Virus Becoming Another Infectious Disease Nightmare
February 19, 2021
https://impakter.com/post-covid-prevent-nipah-virus-infectious-disease-nightmare/
——————————-
Malaysian Researchers Trace Nipah Virus Outbreak to Bats
28 Jul 2000
https://science.sciencemag.org/content/289/5479/518.summary
——————————–
Ebola, Nipah and now COVID-19 — why bats transmit so many deadly viruses
3-12-2020
According to some studies, bats were the original hosts of the novel coronavirus strain that has claimed over 4,000 lives worldwide.
New Delhi: While scientists have yet to irrefutably determine the source of the deadly novel coronavirus strain, which has claimed over 4,000 lives worldwide, some studies indicate that the natural host of COVID-19 are bats.
While bats were not sold at the market in Wuhan, the Chinese region where the virus originated, it is believed that they infected live chickens and animals there.
https://theprint.in/science/ebola-nipah-and-now-covid-19-why-bats-transmit-so-many-deadly-viruses/379853/
——————————–
Scientists discovered a deadly hemorrhagic fever similar to Ebola that can spread person-to-person in Bolivia
11/16/2020
Chapare virus causes a hemorrhagic fever similar to Ebola.
The disease was last observed in Bolivia in 2004 and appeared again in 2019.
Healthcare workers caught the virus by coming into contact with infected bodily fluids.
Experts say data-sharing allowed them to develop a test very quickly. They said Chapare is not a pandemic threat.
https://www.msn.com/en-us/health/medical/scientists-discovered-a-deadly-hemorrhagic-fever-similar-to-ebola-that-can-spread-person-to-person-in-bolivia/ar-BB1b4oxQ
——————————–
Rare and dangerous ‘Chapare’ virus found in Bolivia; can spread from person to person
Nov 18, 2020
The Chapare virus, named after the region where it was first observed, was discovered due to the ongoing efforts of scientists around the globe to avert future pandemics like COVID-19
https://www.businesstoday.in/latest/trends/story/chapare-virus-fatal-new-ebola-like-virus-emerges-in-bolivia-can-spread-among-people-say-scientists-279040-2020-11-18
——————————–
Monitoring the Deadly Nipah Virus
Program Info
Nipah virus first emerged in Malaysia in 1998, infecting 265 people and resulting in more than 100 deaths in a single outbreak. This lethal virus was first identified in pigs and pig farmers. It spread quickly as infected pigs were bought and sold, eventually infecting abattoir workers in Singapore. The Malaysian government was forced to stop pig exports and culled almost half of its pig population, costing the industry millions of dollars. This severe action halted the spread of this newly discovered disease but many questions remained to its origins and pathways.
Joint investigations suggest a complex ecology for Nipah virus and researchers identified fruit bats as the wildlife reservoir of the virus. In 2001, an outbreak of Nipah-like virus erupted in Bangladesh resulting in many human fatalities. Bangladesh has experienced outbreaks every year since then and EcoHealth Alliance scientists fear the virus has pandemic potential since it is easily transmissible among wildlife, people and livestock. The most recent outbreak that occurred in Bangladesh in 2004 had a case fatality rate of nearly 80 percent – a staggering figure.
Our work, in cooperation with local and international partners, involves catching and testing fruit bats, identifying exposure risk to humans, using mathematical models to understand the dynamics of transmission, and predicting how likely it is to become pandemic. EcoHealth Alliance’s goal is to find methods to prevent transmission while continuing to work on ways to protect fruit bats in their natural habitat.
https://www.ecohealthalliance.org/program/monitoring-the-deadly-nipah-virus
——————————-
Bats’ unique immune systems make them stealthy viral reservoirs
These ecologically important animals have been at the center of many major viral outbreaks, and we are beginning to understand why
In 2001, Nipah virus emerged in India, causing an estimated 66 cases and 45 deaths after people unknowingly drank contaminated raw date palm sap. In 2002, SARS (a coronavirus termed Severe Acute Respiratory Syndrome) arose in Asia, resulting in 8,000 infections and almost 800 deaths globally. And from 2014-2016, the Ebola virus outbreak shocked the world, sickening over 28,000 people and killing over 11,000 across Africa.
https://massivesci.com/articles/bat-immune-systems-ncov-sars-nipah-mers-ebola-coronavirus/
——————————–
Asymptomatic infection of marburg virus reservoir bats is explained by a strategy CORD-Papers
2021-06-28
https://covid19-data.nist.gov/pid/rest/local/paper/asymptomatic_infection_of_marburg_virus_reservoir_bats_is_explained_by_a_strategy
——————————–
After Ebola and Covid, possible case of the virulent Marburg virus
August 7, 2021
Guinea announced on Friday “a probable case” of Marburg virus haemorrhagic fever. She is still struggling with Covid-19 and is just emerging from a new Ebola fever epidemic…
https://www.archyde.com/after-ebola-and-covid-possible-case-of-the-virulent-marburg-virus/
——————————–
What’s more deadly than coronavirus?
Feb 20, 2020
Here’s a brief history of the Marburg virus, which was more deadly than the new 2019 virus.
https://thehill.com/changing-america/well-being/prevention-cures/483907-before-the-new-coronavirus-marburg-virus-gave-us
——————————–
Scientists identify new mechanisms that offer protection against Marburg infection
Apr 22 2020
https://www.news-medical.net/news/20200422/Scientists-identify-new-mechanisms-that-offer-protection-against-Marburg-infection.aspx
——————————–
Antibody finding raises hopes for Marburg, COVID-19 treatments
Apr. 30, 2020
https://news.vumc.org/2020/04/30/antibody-marburg-covid-19/
——————————–
Lectin Affinity Plasmapheresis for Middle East Respiratory Syndrome-Coronavirus and Marburg Virus Glycoprotein Elimination
2018
https://pubmed.ncbi.nlm.nih.gov/29698959/
——————————–
Monoclonal antibody cures Marburg infection in monkeys
April 5, 2017
NIH-funded groups preparing for next filovirus outbreak.
https://www.nih.gov/news-events/news-releases/monoclonal-antibody-cures-marburg-infection-monkeys
——————————–
Could Intravenous Immunoglobulin Collected from Recovered Coronavirus Patients Protect against COVID-19 and Strengthen the Immune System of New Patients?
2020 Mar
https://www.ncbi.nlm.nih.gov/pubmed/32218340
———————————
Antiviral inhibitor DDX42 shows broad activity against Chikungunya, HIV and SARS-CoV-2
Oct 29 2020
https://www.news-medical.net/news/20201029/Antiviral-inhibitor-DDX42-shows-broad-activity-against-Chikungunya-HIV-and-SARS-CoV-2.aspx
———————————
COVID-19 Masquerading as Chikungunya Fever
2020
Abstract
The COVID-19 outbreak is an unprecedented global public health challenge. It has a myriad of clinical presentations including fever, cough, vomiting, and diarrhea. Here, we present a unique case of COVID-19, with an atypical presentation of arthralgias and false-positive results for the chikungunya virus. By highlighting the importance of this rare association, we want physicians to be vigilant in the time of this pandemic and to have a high suspicion for this novel disease.
https://thescipub.com/abstract/10.3844/ajidsp.2020.73.76
———————————
Chikungunya fever and COVID-19: Oral ulcers are a common feature
November 2020
https://onlinelibrary.wiley.com/doi/10.1111/odi.13717
———————————
COVID-19, Chikungunya, Dengue and Zika Diseases: An Analytical Platform Based on MALDI-TOF MS, IR Spectroscopy and RT-qPCR for Accurate Diagnosis and Accelerate Epidemics Control
2021
https://pubmed.ncbi.nlm.nih.gov/33808104/
———————————
RESEARCH HIGHLIGHT: Revisiting old antiviral strategies in better, safer ways to fight emerging viruses like Zika, Chikungunya, and Dengue
January 10, 2018
https://stage.systembio.com/research-highlight-revisiting-old-antiviral-strategies-in-better-safer-ways-to-fight-emerging-viruses-like-zika-chikungunya-and-dengue/
———————————
What chikungunya teaches us about COVID-19
May 19, 2021
https://www.thelancet.com/journals/laninf/article/PIIS1473-3099(21)00272-3/fulltext
———————————
Concerns about COVID-19 and arboviral (chikungunya, dengue, zika) concurrent outbreaks
2020
https://www.scielo.br/j/bjid/a/KYW6KZLDyBB64Rp7nmkwSgb/?lang=en
———————————
Pune: Marked rise in chikungunya cases as focus stays on Cov…
December 6, 2020
https://timesofindia.indiatimes.com/city/pune/marked-rise-in-chikungunya-cases-as-focus-stays-on-ncov/articleshow/79586218.cms
———————————
Coronavirus is causing hives and 4 other skin reactions — here’s what dermatologists warn you to look for
April 21, 2020
https://www.yahoo.com/lifestyle/coronavirus-hives-skin-reactions-dermatologist-152002908.html
———————————
‘Turbo charged’ vaccines post promising results for debilitating chikungunya virus
7 August 2021
One of the two jabs, developed by Oxford University, uses the same technology as the Oxford/AstraZeneca coronavirus jab
https://www.telegraph.co.uk/global-health/science-and-disease/turbo-charged-vaccines-post-promising-results-debilitating-chikungunya/
———————————
Chikungunya and COVID-19 in Brazil: The danger of an overlapping crises
April 2020
https://pubmed.ncbi.nlm.nih.gov/33749830/
———————————
Ministry: Chikungunya and Covid remain under control
06 October 2020
https://www.phnompenhpost.com/national/ministry-chikungunya-and-covid-remain-under-control
———————————
Coronavirus outbreak: Ebola, Zika, Nipah and other viruses that caused hundreds of deaths
Jan 24, 2020
Here are the major recent outbreaks that have affected lives and economies.
https://www.moneycontrol.com/news/trends/current-affairs-trends/coronavirus-outbreak-ebola-zika-nipah-and-other-viruses-that-caused-hundreds-of-deaths-4851551.html
——————————–
Ebola vaccine success shows how to beat COVID-19
February 15th, 2021
https://www.vaccinestoday.eu/stories/ebola-vaccine-success-shows-how-to-beat-covid-19/
——————————–
Newly Discovered Cellular Pathway Blocks Ebola, COVID-19 Viruses
November 3, 2020
https://jamanetwork.com/journals/jama/fullarticle/2772474
Faced with the urgent need for new antiviral strategies, investigators recently uncovered a surprising pathway that host cells use to protect against diverse viruses, including those that cause Ebola and coronavirus disease 2019 (COVID-19). The findings, published in Science, point to a potential novel treatment target.
The work relied on an innovative screening approach—activating mobile genetic sequences within chromosomes called transposons—to look for genes that can prevent Ebola virus infection. DNA transposons’ attractiveness for biomedical research lies in their adept ability to hop from place to place within the genome. These genetic segments are powerful forces of change, sometimes disrupting and other times augmenting gene expression.
“This platform is novel and different for several reasons: It both activates and inactivates genes in a truly genome-wide way, it is fast and inexpensive, and it can be easily applied to different cell types and organisms,” the study’s senior author, Adam Lacy-Hulbert, PhD, of the Benaroya Research Institute at Virginia Mason and the University of Washington in Seattle, said in an interview. Lacy-Hulbert noted that many conventional screens knock out host genes to identify those that a virus needs to replicate. “We were most interested in turning on genes and finding ones that protected the cell,” he explained.
The strategy uncovered the major histocompatibility complex (MHC) class II transactivator (CIITA) gene as a promising candidate. In human cell lines, the gene induced Ebola virus resistance by activating a second gene’s expression. Indeed, knockout experiments showed that CIITA-mediated resistance required the other gene, CD74.
Both genes play a role in the body’s immune response, where they’re associated with antigen presentation. The new study suggests that their antiviral activity may have preceded their known role in presenting antigens to the immune system.
“I think a major impact is the revelation that a gene (CD74) that was thought only to function in adaptive immunity (helping process MHCII) has a direct function in innate immunity (inhibiting viral entry by blocking fusion),” Duane Wesemann, MD, PhD, an associate professor and immunology faculty member at Harvard Medical School and a researcher at Brigham and Women’s Hospital, noted by email. “This is a nice example of how genes were co-opted for other functions throughout evolution,” added Wesemann, who was not involved with the study.
Human cells express 4 main types, or isoforms, of the CD74 protein, and only 1 of these isoforms, named p41, was critical for viral resistance in the scientists’ experiments. Its expression fully restored Ebola virus resistance in CIITA-expressing CD74-knockout cells. Even in the absence of the CIITA gene, p41 expression induced antiviral activity.
The p41 isoform blocks cellular enzymes, or proteases, called cathepsins that Ebola viruses hijack for cell entry. Specifically, it inhibits cathepsin proteases from altering certain proteins on the Ebola virus coat. By doing so, it prevents the virus from fusing with and infecting the host cell. In fact, disrupting the isoform’s cathepsin binding site completely inhibited its antiviral activity in the study.
“The pathway we found interferes with an early stage of viral entry that is common to many viruses: the processing of the viral coat protein by host cell proteases,” Lacy-Hulbert said.
In recent months, he and his colleagues have been working to help understand, treat, and prevent COVID-19. Knowing that diverse viruses rely on cathepsins and other proteases, they next tested p41’s potential against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Expressing p41 in cells blocked the virus’s cathepsin-dependent entry pathway and completely abolished signs of cell destruction during viral exposure.
The strategy worked for inhibiting a range of Ebolavirus species, including Sudan, Zaire, and Reston, the distantly related Marburg virus, as well as a bat virus related to SARS-CoV-2 and viruses engineered to express SARS-CoV spike (S) proteins. Human coronaviruses have S proteins in common. They are indispensable for receptor recognition, viral attachment, and host cell entry and represent promising targets for COVID-19 vaccine and therapeutic research.
Anna Bruchez, PhD, the study’s lead author and a pathology instructor at Case Western Reserve University, stressed the findings’ broader implications against known and emerging pathogens. “Many viruses use cathepsin proteases to help them infect cells, and cells can upregulate CIITA and CD74 during an immune response, so it will be interesting to see if expression levels of these genes are associated with differential clinical outcomes following infection,” she said in an interview.
Moving forward, the findings may inspire new treatments that inhibit multiple virus types, including unknown future pathogens. “Ideally, we need to find a drug that can mimic the ability of CD74 to block viral processing,” Lacy-Hulbert said.
——————————–
Ebola, COVID-19, and emerging infectious disease: lessons learned and future preparedness
2020
https://pubmed.ncbi.nlm.nih.gov/32740063/
——————————–
Ebola, COVID-19 and Africa: What we expected and what we got
Sep 2020
https://pubmed.ncbi.nlm.nih.gov/32951291/
——————————–
Is it COVID-19 or Ebola?
https://www.cdc.gov/vhf/ebola/pdf/2018/english/Is-It-COVID19-or-Ebola-English-508.pdf
——————————–
New disease as contagious as Covid and fatal like Ebola approaching humanity: Report
January 04, 2021
https://www.thehindubusinessline.com/news/science/new-disease-as-contagious-as-covid-and-fatal-like-ebola-approaching-humanity-report/article33492726.ece
The scientist who cautioned the world about the Ebola virus haswarned that new and potentially fatal viruses are sprouting from African tropical rainforests.
This comes as a woman demonstrated hemorrhagic fever symptoms in a remote town in the Democratic Republic of the Congo. This has made people apprehensive of new deadly pathogens approaching humanity.
According to a CNN report, the woman in Ingende took a test of several diseases, including Ebola, but all results came out negative.
The report speculated that the disease came from some unknown virus source, an “unexpected” pathogen that could spread as rapidly as Covid-19 but has Ebola’s fatality rate of 50 to 90 per cent. However, the patient who showed the symptoms of this unknown virus recuperated from it.
The World Health Organization (WHO) has called this “Disease X”, where “X” stands for unexpected, is hypothetical for now.
——————————–
‘Ebola-Like Hemorrhagic Fever Virus’ Now Spreading In China, mRNA Vaccine Technology Inventor Reveals
Jan 11, 2022
https://www.christianitydaily.com/articles/14560/20220111/ebola-like-hemorrhagic-fever-virus-now-spreading-in-china-mrna-vaccine-technology-inventor-reveals.htm
——————————–
NEW THREAT: Scientists have found a NEW bat virus that’s 94.5% identical to Covid
Mar 12 2021
https://www.the-sun.com/news/2499086/scientists-found-new-bat-virus-identical-to-covid/
Scientists have previously predicted we could face a health emergency or pandemic every five years likely caused by “zoonotic” diseases – when infections jump from animals to humans.
The threat of unknown viruses that can be transmitted to humans and potentially cause widespread epidemics is known as Disease X by the World Health Organization (WHO).
Bats are known hosts to a broad range of viruses that can cause severe disease in humans.
And the latest identified, called RpYN06, has almost entirely the same genetics as SARS-CoV-2 – the coronavirus that causes Covid-19.
Scientists at Shandong First Medical University & Shandong Academy of Medical Sciences in Taian, China, made the discovery.
They said RpYN06 was the closest relative ever found to some of the individual genes in Covid’s makeup.
However, so far, no Covid-like bat viruses have been discovered that have a spike protein the same as SARS-CoV-2.
The spike protein is the structure on the outside of the virus that it uses to bind to human cells.
The team in China looked at the genetics of 411 samples collected from 23 bat species in Yunnan province in China during 2019 and 2020.
Four viruses related to SARS-CoV-2 were discovered, including RpYN06.
“The other three SARS-CoV-2 related coronaviruses were nearly identical in sequence,” the authors wrote in their paper – which has not been published in a journal.
Scientists, led by Weifeng Shi, said “relatives of SARS-CoV-2 circulate in wildlife species in a broad geographic region of Southeast Asia and southern China”.
They said their findings “highlight the remarkable diversity of bat viruses”.
But they warned it is “essential that further surveillance efforts should cover a broader range of wild animals in this region to help track ongoing spillovers of [viruses] from animals to humans”.
It echoes the thoughts of researchers who recently found another coronavirus closely linked with Covid in bats in eastern Thailand.
RacCS203 shares 91.5 per cent of its genetic code with the Covid-causing virus.
And the closest relative to RacCS203 was RmYN02 – 93.7 per cent identical to Covid.
But the scientists from Chulalongkorn University in Bangkok said the viruses are not thought to be able to infect humans as it is unable to bind to cells in the body because of its differences in the spike protein.
They theorise the virus must change its shape in order to effectively infect other species, or humans.
——————————–
New virus found in bats is nearly 95% identical to Covid
12 Mar 2021
https://metro.co.uk/2021/03/12/new-virus-found-in-bats-is-nearly-95-identical-to-covid-14233848/
——————————–
New Ebola-like virus called Mengla discovered in China
January 10, 2019
https://newatlas.com/ebola-like-filovirus-mengla-china/57978/
——————————–
From Nipah to coronavirus, a story of bats battering humans
3-3-2020
Fewer forests means humans are staying nearer to forests than before, and they now have more chances of exposure to bats and their eating and breeding places.
https://www.indiatoday.in/science/story/coronavirus-covid19-nipah-sars-mers-ebola-marburg-hendra-bats-humans-1652083-2020-03-03
——————————–
Promising new antivirals target arboviruses and SARS-CoV-2: Nodosome inhibitors
Nov 10 2020
https://www.news-medical.net/news/20201110/Promising-new-antivirals-target-arboviruses-and-SARS-CoV-2-Nodosome-inhibitors.aspx
——————————–
SARS-CoV-2 and arbovirus infection: a rapid systematic review
2020
https://pubmed.ncbi.nlm.nih.gov/33111923/
——————————–
Arboviruses and COVID-19: the need for a holistic view
2020
In June, the Pan American Health Organization (PAHO) and WHO published their latest epidemiological update on dengue and other arboviruses. They state that, during the first few months of 2020 (epidemiological week 1–21), there were more than 1·6 million reported cases of arboviral disease in the WHO region of the Americas. Most of these (about 97%) were dengue and the rest were chikungunya (>2%) and Zika (<1%). This prevalence is much lower than that reported in the same period in 2019. A major factor in this overall decrease was a large reduction in cases of chikungunya and Zika virus infection, although cases of dengue also decreased.
Of the total cases of dengue, only a very small proportion (0·2%) were classified as severe dengue and the reported number of these cases fell relative to overall cases when compared with the same period in 2019. Although the reported dengue cases were primarily in Brazil, most of the severe cases were reported in Honduras. Most reports of chikungunya and Zika were also from Brazil—the most populous county in the region. The scale of these diseases in 2020 is less than for 2019 but not trivial. In other years, there would be some comfort taken from the decline in cases, but matters are uncertain given the ongoing COVID-19 pandemic.
https://www.thelancet.com/journals/lanmic/article/PIIS2666-5247(20)30101-4/fulltext
——————————–
COVID-19 and its impact on the control of Aedes (Stegomyia) aegypti mosquito and epidemiological surveillance of arbovirus infections
2021
https://pubmed.ncbi.nlm.nih.gov/34270541/
——————————–
Arbovirus infections of the nervous system: Current trends and future threats
January 27, 2015
https://n.neurology.org/content/84/4/421
——————————–
Arboviruses and COVID-19: the need for a holistic view
2020
https://www.thelancet.com/journals/lanmic/article/PIIS2666-5247(20)30101-4/fulltext
——————————–
Antibody finding raises hopes for Marburg, COVID-19 treatments
April 30, 2020
https://www.vumc.org/crowe-lab/news/antibody-finding-raises-hopes-marburg-covid-19-treatments
Marburg is a distant, more lethal cousin of the RNA virus that causes COVID-19. An outbreak of Marburg hemorrhagic fever in Angola in 2004-2005 killed 90% of the approximately 250 people it infected.
Now researchers at the University of Texas Medical Branch in Galveston and Vanderbilt University Medical Center led by Alexander Bukreyev, PhD, and James Crowe, MD, have isolated non-neutralizing monoclonal antibodies from a Marburg survivor that protect animals from being killed by the virus.
The antibodies bound to the outer envelope protein of the Marburg virus. And while they didn’t kill the virus directly, they recruited other immune cells and antibodies that rapidly cleared the infection.
These findings, reported last week in the journal Cell Host & Microbe, suggest that the unique biological properties of these antibodies make them attractive candidates for therapeutic, monoclonal antibody “cocktails” against Marburg infection, the researchers concluded.
——————————–
Researchers uncover mechanisms of protective antibody response during Marburg infection
April 21, 2020
Summary:
A detailed study of the monoclonal antibodies from a person who survived a Marburg infection led researchers to identify novel mechanisms that contribute protection against the disease.
https://www.sciencedaily.com/releases/2020/04/200421134400.htm
——————————–
Fruit bats carry deadly Marburg virus
September 1, 2007
https://www.newscientist.com/article/dn12567-fruit-bats-carry-deadly-marburg-virus/
——————————–
Coronavirus: US authorises use of anti-viral drug Remdesivir
May 2020
https://www.bbc.com/news/world-us-canada-52511270
The US’s Food and Drug Administration (FDA) has authorised emergency use of the Ebola drug remdesivir for treating the coronavirus.
The authorisation means the anti-viral drug can now be used on people who are hospitalised with severe Covid-19.
A recent clinical trial showed the drug helped shorten the recovery time for people who were seriously ill.
However, it did not significantly improve survival rates.
——————————–
Dr. Ardis: The Medical Industry Is Responsible For ‘Covid’ Deaths, Not Virus
Sep 20, 2021
https://banned.video/watch?id=614916e2e20eb000d7b8d41e
——————————–
Hospital Pharmacies Pushing for Use of New Vancomycin Guidelines Despite COVID-19
August 10, 2020
https://www.pharmacytimes.com/view/hospital-pharmacies-pushing-for-use-of-new-vancomycin-guidelines-despite-covid-19
In March 2020, the new guidelines for vancomycin dosing were released, with the major changes including the transition from a trough-based monitoring process to area under the curve (AUC)-based dosing strategies.
A survey conducted between March 30 and April 6 by InsightRx found that implementing the new vancomycin guidelines into the pharmacy community is as important as coronavirus disease 2019 (COVID-19) in the health care system. Further, a second survey was conducted by Sage Growth Partners in April 2020 to assess pharmacists’ sentiments regarding newly released vancomycin dosing guidelines and related precision dosing topics.
In the April survey, 126 pharmacists were questioned on topics that included their plans to adopt the new vancomycin guidelines and related precision dosing issues. Nearly half of the participants were directors of pharmacy, with 92% working in short-term acute care hospitals.
Some of the key findings of the survey include 52% of respondents indicating that accurately dosing drugs with a narrow therapeutic window, such as vancomycin, is moderately challenging, whereas 33% said it is very or extremely challenging.
As for the most prevalent approaches for estimating vancomycin doses, 55% of respondents base their estimates on professional judgement compared with 49% who base theirs on manual calculation methods.
Although 98% of respondents said that the new vancomycin guidelines are important for patient safety, 86% of respondents are still using trough-based dosing, which the new consensus guidelines recommend be discontinued.
Despite the appreciation of the benefits of the new vancomycin guidelines, only 31% of respondents plan to adopt the new recommendations, whereas 60% are uncertain whether they will do so. Two of the major reasons for not shifting to AUC-guided dosing include not having the budget for the proper software and being content with current vancomycin dosing methods.2
——————————–
Repurposing FDA-approved drugs to fight COVID-19 using in silico methods: Targeting SARS-CoV-2 RdRp enzyme and host cell receptors (ACE2, CD147) through virtual screening and molecular dynamic simulations
2021 Feb 25
https://pubmed.ncbi.nlm.nih.gov/33649734/
Abstract
Background: Different approaches have been proved effective for combating the COVID-19 pandemic. Accordingly, in silico drug repurposing strategy, has been highly regarded as an accurate computational tool to achieve fast and reliable results. Considering SARS-CoV-2’s structural proteins and their interaction the host’s cell-specific receptors, this study investigated a drug repurposing strategy aiming to screen compatible inhibitors of FDA-approved drugs against viral entry receptors (ACE2 and CD147) and integral enzyme of the viral polymerase (RdRp).
Methods: The study screened the FDA-approved drugs against ACE2, CD147, and RDRP by virtual screening and molecular dynamics (MD) simulation.
Results: The results of this study indicated that five drugs with ACE2, four drugs with RDRP, and seven drugs with CD147 achieved the most favorable free binding energy (ΔG < -10). This study selected these drugs for MD simulation investigation whose results demonstrated that ledipasvir with ACE2, estradiol benzoate with CD147, and vancomycin with RDRP represented the most favorable ΔG. Also, paritaprevir and vancomycin have good binding energy with both targets (ACE2 and RdRp).
Conclusions: Ledipasvir, estradiol benzoate, and vancomycin and paritaprevir are potentially suitable candidates for further investigation as possible treatments of COVID-19 and novel drug development.
——————————–
Impact of COVID-19 on pneumonia-focused antibiotic use at an academic medical center
23 July 2020
https://www.cambridge.org/core/journals/infection-control-and-hospital-epidemiology/article/impact-of-covid19-on-pneumoniafocused-antibiotic-use-at-an-academic-medical-center/497EE16254834FA2259D8FB096CA5B49
——————————–
The use of antibiotics in COVID-19 management: a rapid review of national treatment guidelines in 10 African countries
23 June 2021
https://tropmedhealth.biomedcentral.com/articles/10.1186/s41182-021-00344-w
——————————–
Antiviral Drugs That Are Approved or Under Evaluation for the Treatment of COVID-19
July 8, 2021
Remdesivir
Ivermectin
Nitazoxanide
Hydroxychloroquine or Chloroquine and/or Azithromycin
Lopinavir/Ritonavir and Other HIV Protease Inhibitors
Antiviral Therapy
https://www.covid19treatmentguidelines.nih.gov/therapies/antiviral-therapy/summary-recommendations/
——————————–
Neuroleptic malignant syndrome in patients with COVID-19
2020 May 22
https://pubmed.ncbi.nlm.nih.gov/32473756/
Abstract
We report the first two cases of Coronavirus Disease 2019 (COVID-19) who were receiving intensive care including favipiravir, and were clinically diagnosed with neuroleptic malignant syndrome (NMS) to focus attention on NMS in COVID-19 management. Case 1: A 46-year-old-man with acute respiratory distress syndrome (ARDS) caused by COVID-19 infection was being administered favipiravir. Fentanyl, propofol, and rocuronium were also given. On day 3, midazolam administration was initiated for deep sedation. On day 5, his high body temperature increased to 41.2 °C, creatine kinase level elevated, and he developed tachycardia, tachypnea, altered consciousness, and diaphoresis. NMS was suspected, and supportive therapy was initiated. High-grade fever persisted for 4 days and subsided on day 9. Case 2: A 44-year-old-man with ARDS caused by COVID-19 infection was being treated with favipiravir. On day 5, risperidone was started for delirium. On day 7, his body temperature suddenly increased to 40.8 °C, his CK level elevated, and he developed tachycardia, tachypnea, altered consciousness, and diaphoresis. NMS diagnosis was confirmed, and both, favipiravir and risperidone were discontinued on day 8. On the same day, his CK levels decreased, and his body temperature normalized on day 9. Patients with COVID-19 infection frequently require deep sedation and develop delirium; therefore, more attention should be paid to the development of NMS in patients who are being administered such causative agents. The mechanism underlying the occurrence of NMS in COVID-19 patients treated with favipiravir remains unknown. Therefore, careful consideration of NMS development is necessary in the management of COVID-19 patients.
——————————–
Midazolam Used to Kill Countless Non-Covid Patients so They Could Be Labelled Coronavirus Deaths
September 15, 2021
http://stateofthenation.co/?p=84012
——————————–
Hospitals murdering COVID-19 patients by forcing them to take deadly drugs like remdesivir and midazolam – Clay Clark on Brighteon.TV
September 29, 2021
https://www.planet-today.com/2021/09/hospitals-murdering-covid-19-patients.html#gsc.tab=0
——————————–
Hospitals murdering COVID-19 patients by forcing them to take deadly drugs like remdesivir and midazolam – Clay Clark on Brighteon.TV
September 30, 2021
https://dreddymd.com/2021/09/30/hospitals-murdering-covid-19-patients-by-forcing-them-to-take-poisonous-drugs/
——————————–
Supplies of sedative used for COVID-19 patients diverted from France to avoid potential shortages
12 February 2021
Exclusive: Accord Healthcare has told The Pharmaceutical Journal that “some French label stock” of midazolam is now being sold into UK wholesalers.
Supplies of the sedative midazolam have been diverted from France as a “precaution” to mitigate potential shortages in the NHS caused by COVID-19, the Department of Health and Social Care (DHSC) has told The Pharmaceutical Journal.
A spokesperson from Accord Healthcare, one of five manufacturers of the drug, told The Pharmaceutical Journal that it had to gain regulatory approval to sell French-labelled supplies of midazolam injection to the NHS, after having already sold two years’ worth of stock to UK wholesalers “at the request of the NHS” in March 2020.
The DHSC said the request for extra stock was part of “national efforts to respond to the coronavirus outbreak”, which included precautions “to reduce the likelihood of future shortages”.
Midazolam is listed by the Royal College of Anaesthetists as a “first-line” sedative in the management of COVID-19 patients, and warns in guidance published on 2 April 2020 that it “may be subject to demand pressure”.
Matt Hancock, the UK health secretary, told the House of Commons Health and Social Care Select Committee on 17 April 2020 that intensive therapy unit medicines — including midazolam — are part of “a delicate supply chain” as they “are made in a relatively small number of factories around the world”.
While the DHSC confirmed that midazolam is still available to both primary and secondary care, it added that some suppliers of the sedative had limited or no stock availability.
A spokesperson from Accord Healthcare told The Pharmaceutical Journal on 11 May 2020 that it was out of stock of midazolam injection after the NHS requested it “place all of its stock of midazolam — equivalent to around two year’s forecasted supply — into its wholesale partners”, even though the manufacturer “does not currently have any NHS contracts in England” to supply the drug.
“As a result of the NHS request [in March 2020], we are subsequently out of stock,” said Peter Kelly, managing director of Accord Healthcare.
However, he added that the Medicines and Healthcare products Regulatory Agency (MHRA) had given the manufacturer approval “for some French label stock — another 22,000 packs — to be sold into the NHS and [we] are currently waiting for the MHRA’s direction on where to place the stock”.
The manufacturer said the French stock only includes midazolam at the strength of 1mg/mL in 5mL, while the initial supply in March 2020 contained a variety of four different strengths.
A spokesperson for the DHSC said it was “working closely with industry, the NHS and others in the supply chain to help ensure patients can access the medicines they need and precautions are in place to reduce the likelihood of future shortages”.
The DHSC confirmed that its request for additional midazolam stock from Accord Healthcare was one of these precautions.
“As part of our national efforts to respond to the coronavirus outbreak, we are doing everything we can to ensure patients continue to access safe and effective medicines,” they added.
https://pharmaceutical-journal.com/article/news/supplies-of-sedative-used-for-covid-19-patients-diverted-from-france-to-avoid-potential-shortages
——————————–
Midazolam
https://en.wikipedia.org/wiki/Midazolam#Use_in_executions
Midazolam, sold under the brand name Versed, among others, is a benzodiazepine medication used for anesthesia, procedural sedation, trouble sleeping, and severe agitation. It works by inducing sleepiness, decreasing anxiety, and causing a loss of ability to create new memories. It is also useful for the treatment of seizures. Midazolam can be given by mouth, intravenously, or injection into a muscle, by spraying into the nose, or through the cheek. When given intravenously, it typically begins working within five minutes; when injected into a muscle, it can take fifteen minutes to begin working. Effects last for between one and six hours.
Side effects can include a decrease in efforts to breathe, low blood pressure, and sleepiness. Tolerance to its effects and withdrawal syndrome may occur following long-term use. Paradoxical effects, such as increased activity, can occur especially in children and older people. There is evidence of risk when used during pregnancy but no evidence of harm with a single dose during breastfeeding. It belongs to the benzodiazepine class of drugs and works by increasing the activity of the GABA neurotransmitter in the brain.
Midazolam was patented in 1974 and came into medical use in 1982. It is on the World Health Organization’s List of Essential Medicines. Midazolam is available as a generic medication. In many countries, it is a controlled substance.
Contents
1 Medical uses
1.1 Seizures
1.2 Procedural sedation
1.3 Agitation
1.4 End of life care
2 Contraindications
3 Side effects
3.1 Pregnancy and breastfeeding
3.2 Elderly
3.3 Tolerance, dependence, and withdrawal
3.4 Overdose
3.5 Detection in body fluids
3.6 Interactions
4 Pharmacology
5 History
6 Society and culture
6.1 Cost
6.2 Availability
6.3 Legal status
6.4 Marketing authorization
6.5 Use in executions
6.5.1 Notable incidents
6.5.2 Legal challenges
——————————–
Doctors: Execution drugs could help COVID-19 patients
April 21, 2020
https://abcnews.go.com/US/wireStory/doctors-execution-drugs-covid-19-patients-70264297
A group of medical professionals is asking death penalty states for medications used both for lethal injections and to help coronavirus patients who are on ventilators
HOUSTON — Secrecy surrounding executions could hinder efforts by a group of medical professionals who are asking the nation’s death penalty states for medications used in lethal injections so that they can go to coronavirus patients who are on ventilators, according to a death penalty expert and a doctor who’s behind the request.
In a letter sent this month to corrections departments, a group of seven pharmacists, public health experts, and intensive care unit doctors asked states with the death penalty to release any stockpiles they might have of execution drugs to health care facilities.
“Your stockpile could save the lives of hundreds of people; though this may be a small fraction of the total anticipated deaths, it is a central ethical directive that medicine values every life,” according to the letter.
But it’s unclear what drugs the states may have, as they have tended to release information about execution protocols and drug supplies only through open records requests or lawsuits. Only one state, Wyoming, responded directly to the letter, and it indicated it doesn’t have the drugs in question.
“I’m not trying to comment on the rightness or wrongness of capital punishment,” said Dr. Joel Zivot, one of the medical professionals who signed the letter. “I’m asking now as a bedside clinician caring for patients, please help me.”
For most people, the new coronavirus causes mild or moderate symptoms, such as fever and cough that clear up in two to three weeks. But for some, it can cause severe illness, requiring them to be placed to ventilators to help them breathe.
Many medications used to sedate and immobilize people put on ventilators and to treat their pain are the same drugs that states use to put inmates to death. Demand for such drugs surged 73% in March.
Twenty-five states have the death penalty, while three have moratoriums on capital punishment.
While some states contacted by The Associated Press, including Alabama and Florida, didn’t respond to inquiries about the letter, others, including Arkansas, Texas and Utah, limited their comment to mainly saying they don’t have the medications in question. Tennessee wouldn’t confirm whether it has the drugs and indicated it has no plans to give any medications to a hospital. Oklahoma said it hadn’t received any requests for such medications from state hospitals.
States may be hesitant to turn over their drugs because they have had problems securing them as many pharmaceutical companies oppose their use in executions, said Robert Dunham, executive director of the Death Penalty Information Center.
Since 2011, 13 states have enacted new statutes that conceal information about the execution process, according to the Death Penalty Information Center, which takes no position on capital punishment but has criticized the way states carry out executions.
Drugs being requested include the sedative midazolam, the paralytic vecuronium bromide and the opioid fentanyl. They’re needed because putting a patient on a ventilator “with no drugs … would be torture,” said Zivot, an associate professor of anesthesiology and surgery at Emory University in Atlanta who has studied medicine’s role in capital punishment.
The tense debate over the supply of execution drugs was highlighted in a 2018 lawsuit that several pharmaceutical companies filed against Nevada over accusations that it illegally obtained its inventory.
In a court brief, 15 states, including Florida, Oklahoma and Texas, called the lawsuit part of the “guerrilla warfare being waged by antideath-penalty activists and criminal defense attorneys to stop lawful executions.”
The lawsuit was dismissed this month after Nevada agreed to return its supplies to the companies, leaving the state without any drugs to carry out executions.
Pharmaceutical companies have long warned that states’ use of these medications for executions could result in shortages, Dunham said.
“Some of the responses over the past several years had been, ‘That’s chicken little saying the sky is falling,’” Dunham said. “But with COVID-19, the sky has fallen.”
——————————–
FDA Documents Show Pfizer Secretly Added Heart Attack Drug to Children’s COVID Vaccines
Nov 10, 2021
https://banned.video/watch?id=618c7b9111848a41725b16e2
——————————–
Analgesia and Sedation Strategies in COVID-19 Patients
March 9, 2021
https://www.uspharmacist.com/article/analgesia-and-sedation-strategies-in-covid19-patients
Sedative Options in COVID-19
Dexmedetomidine would be a viable option in patients able to tolerate light sedation given its opioid-sparing effects, which would help mitigate ongoing shortages. The side-effect profiles of both agents require careful assessment in patients with COVID-19. Propofol can cause hypertriglyceridemia as it is a 10% fat emulsion–containing product; triglycerides should be trended while patients remain on propofol.12 Once triglycerides are greater than 500 mg/dL, nonpropofol sedation strategies should be considered. However, patients with COVID-19 may present with a secondary hemophagocytic lymphohistocytosis (HLH)-type picture with severe hypertriglyceridemia.13 HLH is the result of excessive immune activation and inflammation due to the absence of downregulated activated macrophages and lymphocytes, leading to tissue destruction and high mortality rates. Thus, triglycerides need to be more carefully monitored in this patient population, and clinicians should consider a lower threshold for transitioning to a nonpropofol sedation strategy.
Benzodiazepines are less commonly utilized for sedation given the concern for increased risk of delirium and ICU LOS.5 However, with ongoing national shortages and the potential need for deeper sedation, benzodiazepines utilization has become more frequent. Lorazepam continuous infusions are not typically employed in the ICU population due to the propylene glycol additive, which can lead to a high anion gap metabolic acidosis and renal failure. If a benzodiazepine continuous infusion is needed, midazolam is more commonly given. Midazolam may lead to prolonged sedation when used for long periods of time and will accumulate in patients with renal or hepatic dysfunction, heart failure, or obesity. Providers must be cognizant of midazolam’s active metabolite and potential for prolonged sedation even when a continuous infusion has been discontinued.
As with analgesia, drug shortages have forced clinicians to consider alternative sedation strategies. Likely one of the most effective and practical ways to limit continuous infusions is by utilizing oral dosage forms or intermittent doses of sedatives. For example, agents such as diazepam or lorazepam could be used intermittently to minimize continuous infusion–sedation requirements. Clonidine, an alpha-2 agonist, has some data for use as sedation in an ICU setting and might be appropriate to consider in hemodynamically stable patients.
Ketamine is an agent that has a variety of uses in critical illness, including acute pain management, postoperative analgesia, refractory status epilepticus, and adjunctive sedation.7,16 Given its analgesic properties, it may also be opioid sparing, which would also be beneficial in the setting of drug shortages.17 There are several considerations for using ketamine in this patient population. Although ketamine is associated with hypertension and tachycardia, it may also decrease myocardial function in some subsets of critically ill patients, including those with septic shock.18 Ketamine may also be considered for usage in targeting deep sedation; however, logistical concerns then arise. When infused continuously at higher doses, the limitation starts to become the amount of volume being infused, particularly if patients are unable to keep up with their fluid balance. Liberal fluid management strategies in patients with acute lung injury may be associated with increased ventilator days, days with central nervous system failure, and days in the ICU, so fluid balance is a paramount concern in these patients.
Volatile gases, including desflurane, isoflurane, and sevoflurane, have also garnered some interest as potential contingency sedation options. These agents are typically used as general anesthesia in patients undergoing surgery, and there is very limited evidence for their usage in the ICU, especially for longer periods of time (>24-48 hours).20-22 Other limitations include the need for special-delivery devices and lack of experience with these agents outside of the operating room. Although uncommon, clinicians should be aware of the potential for patients developing malignant hyperthermia with these inhaled anesthetics.
——————————–
26% of those prescribed Remdesivir for COVID died, according to Medicare database
Sep 29, 2021
https://www.lifesitenews.com/news/26-of-those-prescribed-remdesivir-for-covid-died-according-to-medicare-database/
——————————–
Remdesivir against COVID-19 and Other Viral Diseases
2020 Dec 16
Abstract
Patients and physicians worldwide are facing tremendous health care hazards that are caused by the ongoing severe acute respiratory distress syndrome coronavirus 2 (SARS-CoV-2) pandemic. Remdesivir (GS-5734) is the first approved treatment for severe coronavirus disease 2019 (COVID-19). It is a novel nucleoside analog with a broad antiviral activity spectrum among RNA viruses, including ebolavirus (EBOV) and the respiratory pathogens Middle East respiratory syndrome coronavirus (MERS-CoV), SARS-CoV, and SARS-CoV-2. First described in 2016, the drug was derived from an antiviral library of small molecules intended to target emerging pathogenic RNA viruses. In vivo, remdesivir showed therapeutic and prophylactic effects in animal models of EBOV, MERS-CoV, SARS-CoV, and SARS-CoV-2 infection. However, the substance failed in a clinical trial on ebolavirus disease (EVD), where it was inferior to investigational monoclonal antibodies in an interim analysis. As there was no placebo control in this study, no conclusions on its efficacy in EVD can be made. In contrast, data from a placebo-controlled trial show beneficial effects for patients with COVID-19. Remdesivir reduces the time to recovery of hospitalized patients who require supplemental oxygen and may have a positive impact on mortality outcomes while having a favorable safety profile. Although this is an important milestone in the fight against COVID-19, approval of this drug will not be sufficient to solve the public health issues caused by the ongoing pandemic. Further scientific efforts are needed to evaluate the full potential of nucleoside analogs as treatment or prophylaxis of viral respiratory infections and to develop effective antivirals that are orally bioavailable.
——————————–
The dangers of remdesivir to treat COVID-19 patients in hospitals – Brighteon.TV
09/15/2021
https://www.dangerousmedicine.com/2021-09-15-unapproved-medication-used-to-treat-covid19-patients.html
——————————–
Protein found on infected cells protects virus from immune system; remdesivir helps prevent hospitalization
September 2021
https://www.msn.com/en-us/health/medical/protein-found-on-infected-cells-protects-virus-from-immune-system-remdesivir-helps-prevent-hospitalization/ar-AAOSTU8
——————————–
Pharmaceutical Scientist Warns of Potential Problems With Remdesivir As COVID-19 Treatment
December 30, 2020
https://scitechdaily.com/pharmaceutical-scientist-warns-of-potential-problems-with-remdesivir-as-covid-19-treatment/
——————————–
Remdesivir caused dangerously low heart rate in COVID-19 patients
Jun 25 2021
https://www.news-medical.net/news/20210625/Remdesivir-caused-dangerously-low-heart-rate-in-COVID-19-patients.aspx
——————————–
Cardiac Adverse Events With Remdesivir in COVID-19 Infection
October 24, 2020
https://www.cureus.com/articles/44072-cardiac-adverse-events-with-remdesivir-in-covid-19-infection
——————————–
Remdesivir may cause liver and kidney injury: ICMR tells states to use it cautiously
July 13, 2020
https://www.thehealthsite.com/news/remdesivir-may-cause-liver-and-kidney-injury-icmr-tells-states-to-use-it-cautiously-757018/
——————————–
Remdesivir in Patients with Acute or Chronic Kidney Disease and COVID-19
2020 Jun 8
https://pubmed.ncbi.nlm.nih.gov/32513665/
——————————–
Sinus Bradycardia Associated with Remdesivir Treatment in COVID-19: A Case Report and Literature Review
2021 Feb 12
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7918811/
——————————–
Doctor Reveals that Remdesivir Was the Real Cause For Many Alleged COVID-19 Maladies
August 7, 2021
Dr. Bryan Ardis makes an astounding revelation. He states that Dr. Fauci pushed the use of Veklury® (remdesivir) as a treatment for COVID-19 knowing that it would be unsafe and ineffective for patients. Veklury® (remdesivir) is a nucleotide analogue RNA polymerase inhibitor. Dr. Ardis reveals that the symptoms of lungs filling with fluid and the other alleged COVID-19 symptoms were actually side effects of kidney poisoning and other organ damage that are known side-effects of Veklury® (remdesivir). Dr. Ardis alleges that the devestating health toll allegedly caused by COVID-19 was actually caused by the NIH recommended treatment of Veklury® (remdesivir).
https://greatmountainpublishing.com/2021/08/07/doctor-reveals-that-remdesivir-was-the-real-cause-for-many-alleged-covid-19-maladies/
——————————–
Sinus Bradycardia Associated with Remdesivir Treatment in COVID-19: A Case Report and Literature Review
2021 Feb 12
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7918811/
——————————–
Real-world risk evaluation of remdesivir in patients with an estimated glomerular filtration rate of less than 30 mL/min
2021 Jun 9
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8344606/
——————————–
Drug-Induced Liver Injury After COVID-19 Vaccine
July 19, 2021
https://www.cureus.com/articles/63877-drug-induced-liver-injury-after-covid-19-vaccine
——————————–
COVID-19 Vaccines in Patients With Chronic Liver Disease
2021 Jun 19
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8214314/
——————————–
Liver injury following SARS-CoV-2 vaccination: A multicenter case series
2021 Jul 31
https://www.ncbi.nlm.nih.gov/labs/pmc/articles/PMC8324396/
——————————–
Drug-Induced Liver Injury After COVID-19 Vaccine
2021 Jul 19
https://www.ncbi.nlm.nih.gov/labs/pmc/articles/PMC8372667/
Abstract
The first case of coronavirus disease 2019 (COVID-19) was reported in December 2019 in China. World Health Organization declared it a pandemic on March 11, 2020. It has caused significant morbidity and mortality worldwide. Persistent symptoms and serious complications are being reported in patients who survived COVID-19 infection, but long-term sequelae are still unknown. Several vaccines against COVID-19 have been approved for emergency use around the globe. These vaccines have excellent safety profiles with few reported side effects. Drug-induced hepatotoxicity is mainly seen with different drugs or chemicals. There are only a few reported cases of hepatotoxicity with vaccines. We present a case of liver injury after administration of the vaccine against the COVID-19 infection.
Introduction
A novel coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-COV-2) causing coronavirus disease 2019 (COVID-19) emerged in December 2019 in Wuhan, China, resulting in an ongoing pandemic [1]. To date, it has caused more than 173 million cases and over 3.7 million death worldwide as per World Health Organization [2]. Although the respiratory system is the most common system affected by this disease, it affects multiple organ manifestations [3]. Despite international efforts to develop treatments for this disease, there are still limited therapeutic options available with remdesivir as the only Food and Drug Administration-approved drug [4]. Given the rapid spread, high morbidity, and mortality worldwide, a coordinated effort led to developing the vaccine in a year of first diagnosed case. Multiple COVID vaccines have been developed at an unprecedented rate. These vaccines have excelled safety and efficacy profiles [5-7]. The most common adverse effects reported with these vaccines included mild effects like pain at the vaccine site, fever, fatigue, headache, arthralgia, myalgia, lymphadenopathy, and severe effects like anaphylactic reaction [8]. Drug-induced hepatotoxicity is a common adverse event seen with prescription and nonprescription drugs [9]. There are few reported hepatotoxicity cases due to vaccines, namely anti-rabies vaccination-induced hepatotoxicity and autoimmune hepatitis due to influenza virus and hepatitis A and B vaccines [10-17]. We report a case of liver injury after receiving the COVID vaccine.
——————————–
Acute liver failure after vaccination against of COVID-19; a case report and review literature
2021 Dec 14
https://pubmed.ncbi.nlm.nih.gov/34926142/
Abstract
Background:
Vaccination against COVID-19 remains as a main root of COVID-19 prevention. Few vaccines have been launched for this purpose recently with different side effects. Thrombotic events have been reported as a rare side effect after ChAdox1nCOV-19 vaccination that may cause death of recipient.
Case presentation:
We report a case of hepatic artery occlusion after the first dose vaccination by ChAdOx1nCov-19. The patient was a health care worker, aged 34-year old. Past medical history was unremarkable and had not used heparin. Over the next couple of days after the vaccination, he reported headache, nausea, and dizziness as well as abdominal pain. His general status and the laboratories studies deteriorate quickly by increasing liver enzymes and severe coagulopathy. Clinically he had presented acute hepatic failure. He had been received blood products, prednisolone pulse along with broad antibiotics without benefit. He died on the sixth day.
Conclusions:
Thrombotic events after vaccination is very rare but can develop in main arteries with lethal outcome. This event may mimic autoimmune thrombosis clinically.
Keywords:
Adverse events, AEs; Amino acid cleaving enzyme, FURIN; COVID-19; Complete Blood count, CBC; Coronavirus disease 2019, COVID-19; Embolism; European Medicines Agency, EMA; Heparin-induced thrombocytopenia, HIT; Intensive-care unit, ICU; Liver; Partial thrombin time, PTT; Platelet factor 4, PF4; Prothrombin time, PT; Trans membrane serine protease 2, TMPRSS2; UK Medicines and other Health products Regulatory Agency, MHRA; Vaccination.
——————————–
New study suggests COVID-19 vaccine turns into DNA in the liver
March 7, 2022
https://fism.tv/new-study-suggests-covid-19-vaccine-turns-into-dna-in-the-liver/
A new peer-reviewed study by Swedish scientists published in the Current Issues in Molecular Biology medical journal suggests that the mRNA from COVID-19 vaccines enter human liver cells, triggering a number of reactions that causes this mRNA to turn into DNA. The studies were done in vitro, meaning that the study was performed inside human HuH-7 liver cells.
The study authors write that the BNT162b2, Pfizer-BioNTech’s COVID-19 vaccine, is able to enter the human liver cell and be reverse transcribed as quickly as six hours post-vaccination. Reverse transcription occurs through an enzyme called reverse transcriptase, which creates double-stranded DNA from an RNA template.
The study also found spike proteins on the surface of the liver cells, which may trigger the immune system and cause autoimmune hepatitis, which has been identified in “case reports on individuals who developed autoimmune hepatitis after BNT162b2 vaccination.” Autoimmune hepatitis is a rare disease where the immune system attacks the liver causing inflammation, or hepatitis, and can lead to cirrhosis and liver failure. In severe cases a liver transplant may be needed.
One theory is that spike proteins circulate in the body after vaccination instead of remaining at the injection site, however some studies are beginning to reveal that the mRNA moves from the injection site and through the bloodstream to organs such as the liver, spleen, adrenal glands, and ovaries.
More studies, including animal studies, are needed to better understand the effects that mRNA vaccines have on the human body. The authors write “at this stage, we do not know if DNA reverse transcribed from BNT162b2 is integrated into the cell genome. Further studies are needed to demonstrate the effect of BNT162b2 on genomic integrity, including whole genome sequencing of cells exposed to BNT162b2, as well as tissues from human subjects who received BNT162b2 vaccination.”
Dr. Peter McCullough, an internist, cardiologist, and epidemiologist tweeted that these findings have “enormous implications of permanent chromosomal change and long-term constitutive spike synthesis driving the pathogenesis of a whole new genre of chronic disease.”
While the study suggests that COVID-19 mRNA vaccines are capable of becoming DNA, this is something that the CDC, FDA, and Pfizer-BioNTech have adamantly denied, saying that reverse transcription is something that would not happen with the vaccines. Pfizer previously said that the vaccine “only presents the body with instructions to build immunity,” because “it does not alter the DNA sequence of a human cell.”
The CDC reiterates this in their “Myths and Facts” sheet, saying that “the genetic material delivered by mRNA vaccines never enters the nucleus of your cells,” adding, “COVID-19 vaccines do not change or interact with your DNA in any way.”
Throughout the duration of the virus experts have been proven wrong on a variety of things they had said were scientific fact, including the strength of natural immunity and vaccine safety and durability. It now appears they could be wrong about the vaccine relationship with DNA as well.
CDC director Rochelle Walensky recently said that unfortunately the CDC did not have the luxury of “making sure every I is dotted, every T is crossed before a decision is made.” She also added that “when you wait for those perfect data to make a decision, it’s too late. We need to sometimes make decisions in the context of areas where the science is gray.”
——————————–
Intracellular Reverse Transcription of Pfizer BioNTech COVID-19 mRNA Vaccine BNT162b2 In Vitro in Human Liver Cell Line
25 February 2022
Abstract
Preclinical studies of COVID-19 mRNA vaccine BNT162b2, developed by Pfizer and BioNTech, showed reversible hepatic effects in animals that received the BNT162b2 injection. Furthermore, a recent study showed that SARS-CoV-2 RNA can be reverse-transcribed and integrated into the genome of human cells. In this study, we investigated the effect of BNT162b2 on the human liver cell line Huh7 in vitro. Huh7 cells were exposed to BNT162b2, and quantitative PCR was performed on RNA extracted from the cells. We detected high levels of BNT162b2 in Huh7 cells and changes in gene expression of long interspersed nuclear element-1 (LINE-1), which is an endogenous reverse transcriptase. Immunohistochemistry using antibody binding to LINE-1 open reading frame-1 RNA-binding protein (ORFp1) on Huh7 cells treated with BNT162b2 indicated increased nucleus distribution of LINE-1. PCR on genomic DNA of Huh7 cells exposed to BNT162b2 amplified the DNA sequence unique to BNT162b2. Our results indicate a fast up-take of BNT162b2 into human liver cell line Huh7, leading to changes in LINE-1 expression and distribution. We also show that BNT162b2 mRNA is reverse transcribed intracellularly into DNA in as fast as 6 h upon BNT162b2 exposure.
——————————–
The exogenous genetic material coding for the dangerous Spike protein is reverse-transcribed into the human genome; possible long-term constitutive expression/synthesis of disease promoting/lethal Spike. [PEER REVIEWED]
February 2022
https://www.reddit.com/r/DebateVaccines/comments/t1eos1/alden_et_al_lund_university_sweden_confirms_one/?utm_source=share&utm_medium=ios_app&utm_name=iossmf
——————————–
Antibody Response to COVID-19 Vaccines in Liver Disease Patients
March 1, 2021
https://clinicaltrials.gov/ct2/show/NCT04775069
——————————–
Autoimmune hepatitis after COVID-19 vaccine – more than a coincidence
2021 Oct 26
Abstract
The COVID-19 pandemic is still raging across the world and vaccination is expected to lead us out of this pandemic. Although the efficacy of the vaccines is beyond doubt, safety still remains a concern. We report a case of a 65-year-old woman who experienced acute severe autoimmune hepatitis two weeks after receiving the first dose of Moderna-COVID-19 vaccine. Serum immunoglobulin G was elevated and antinuclear antibody was positive (1:100, speckled pattern). Liver histology showed a marked expansion of the portal tracts, severe interface hepatitis and multiple confluent foci of lobular necrosis. She started treatment with prednisolone, with a favorable clinical and analytical evolution. Some recent reports have been suggested that COVID-19 vaccination can lead to the development of autoimmune diseases. It is speculated that the vaccine can disturb self-tolerance and trigger autoimmune responses through cross-reactivity with host cells. Therefore, healthcare providers must remain vigilant during mass COVID-19 vaccination.
https://pubmed.ncbi.nlm.nih.gov/34717185/
——————————–
Pfizer Report: Confidential (Japanese)
https://www.docdroid.net/xq0Z8B0/pfizer-report-japanese-government-pdf
——————————–
COVID-19 liver damage may be more common than previously thought
August 12, 2020
Compared with hospitalized patients in China, a new study finds higher rates of abnormal liver tests among patients receiving treatment in hospitals in the United States. Higher levels of liver enzymes seem to be associated with an increased risk of admission to intensive care and death.
https://www.medicalnewstoday.com/articles/covid-19-liver-damage-may-be-more-common-than-previously-thought
——————————–
Covid treatment: Study finds remdesivir effective against key enzyme that causes COVID-19
Jul 26, 2021
COVID treatment: A drug used to treat hepatitis C and other cold-like viruses has shown to be effective by interfering with one of the key enzymes the virus needs to replicate RNA, preventing the virus from multiplying.
https://www.express.co.uk/life-style/health/1467557/covid-treatment-remdesivir-enzymes-preventing-covid-19
——————————–
How The New Coronavirus Compares to Past Zoonotic Outbreaks, in One Simple Chart
1-31-2020
https://www.sciencealert.com/this-chart-shows-how-the-wuhan-virus-compares-to-other-recent-outbreaks
The chart above does not include mosquito-borne viruses like Zika and Dengue fever.
All of these diseases jumped from animals to people
According to experts, the new coronavirus likely originated in bats. More than 75 percent of emerging diseases originate in animals; these are called zoonotic diseases, meaning they can jump from animals to people.
At least 10 outbreaks in the last century have spilled over to humans from animals such as bats, birds, and pigs.
——————————–
Experts think bats are the source of the Wuhan coronavirus. At least 4 pandemics have originated in these animals.
Feb 10, 2020
In the past 45 years, at least three other pandemics (besides SARS) have been traced back to bats. The creatures were the original source of Ebola, which has killed 13,500 people in multiple outbreaks since 1976; Middle Eastern respiratory syndrome, better known as MERS, which can be found in 28 countries; and the Nipah virus, which has a 78% fatality rate.
https://www.businessinsider.com/wuhan-coronavirus-sars-bats-animals-to-humans-2020-1
——————————-
Henipavirus
https://en.wikipedia.org/wiki/Henipavirus
Henipavirus is a genus of RNA viruses in the family Paramyxoviridae, order Mononegavirales containing five established species. Henipaviruses are naturally harboured by pteropid fruit bats (flying foxes) and microbats of several species. Henipaviruses are characterised by long genomes and a wide host range. Their recent emergence as zoonotic pathogens capable of causing illness and death in domestic animals and humans is a cause of concern.
In 2009, RNA sequences of three novel viruses in phylogenetic relationship to known henipaviruses were detected in African straw-colored fruit bats (Eidolon helvum) in Ghana. The finding of these novel henipaviruses outside Australia and Asia indicates that the region of potential endemicity of henipaviruses may be worldwide. These African henipaviruses are slowly being characterised.
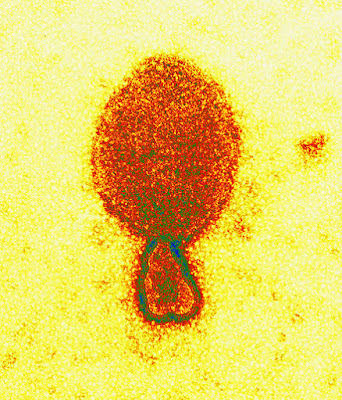
Colored transmission electron micrograph of a Hendra henipavirus virion (ca. 300 nm length)
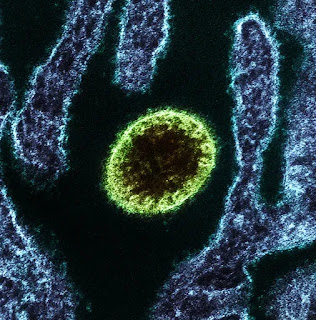
(Nipah)

False-color electron micrograph showing a Nipah virus particle (purple) by an infected Vero cell (brown)
——————————-
Possible role of the Nipah virus V protein in the regulation of the interferon beta induction by interacting with UBX domain-containing protein1
16 May 2018
https://www.nature.com/articles/s41598-018-25815-9
Abstract
Nipah virus (NiV) is a highly pathogenic paramyxovirus that causes lethal encephalitis in humans. We previously reported that the V protein, one of the three accessory proteins encoded by the P gene, is one of the key determinants of the pathogenesis of NiV in a hamster infection model. Satterfield B.A. et al. have also revealed that V protein is required for the pathogenicity of henipavirus in a ferret infection model. However, the complete functions of NiV V have not been clarified. In this study, we identified UBX domain-containing protein 1 (UBXN1), a negative regulator of RIG-I-like receptor signaling, as a host protein that interacts with NiV V. NiV V interacted with the UBX domain of UBXN1 via its proximal zinc-finger motif in the C-terminal domain. NiV V increased the level of UBXN1 protein by suppressing its proteolysis. Furthermore, NiV V suppressed RIG-I and MDA5-dependent interferon signaling by stabilizing UBXN1 and increasing the interaction between MAVS and UBXN1 in addition to directly interrupting the activation of MDA5. Our results suggest a novel molecular mechanism by which the induction of interferon is potentially suppressed by NiV V protein via UBXN1.
———————
Distinct and overlapping roles of Nipah virus P gene products in modulating the human endothelial cell antiviral response.
2012
https://www.ncbi.nlm.nih.gov/pubmed/23094089
———————
———————
Vaccine Development for Nipah Virus Infection in Pigs
2-4-2019
https://www.frontiersin.org/articles/10.3389/fvets.2019.00016/full
Nipah virus is an Emerging Pathogen With the Potential For Pandemic
Nipah virus (NiV) is an enveloped, single stranded, negative sense RNA paramyxovirus, genus Henipavirus. The natural hosts and wildlife reservoirs of NiV are Old World fruit bats of the genus Pteropus. Both Nipah and the related Hendra virus possess a number of features that distinguish them from other paramyxoviruses. Of particular note is their broad host range which is facilitated by the use of the evolutionary conserved ephrin-B2 and –B3 as cellular receptors. The NiV attachment glycoprotein (G) is responsible for binding to ephrin-B2/-B3 (3). Following receptor binding, the G protein dissociates from the fusion (F) protein. Subsequently, the F protein undergoes a series of conformational changes which in turn initiates fusion of the viral and host membrane allowing entry. During viral replication, the F protein is synthesized and cleaved into fusion active F1 and F2 subunits. These subunits are subsequently transported back to the cell surface to be incorporated into budding virions, or facilitate fusion between infected and adjacent uninfected cells. This cell-to-cell fusion results in the formation of multinucleated cells called syncytia, and greatly influences the cyopathogenicity of NiV as it allows spread of the virus, even in the absence of viral budding.
NiV infection is currently classed as a stage III zoonotic disease, meaning it can spill over to humans and cause limited outbreaks of person-to-person transmission. NiV outbreaks have been recognized yearly in Bangladesh since 2001 as well as occasional outbreaks in neighboring India (Figure 1). These outbreaks have been characterized by person-to-person transmission and the death of over 70% of infected people. In May 2018, the first ever outbreak in southern India was reported. A total of 19 NiV cases, of which 17 resulted in death, were reported in the state of Kerala. Pteropus giganteus bats from areas around the index case in Kozhikode, Kerala, were tested at the National High Security Animal Diseases Laboratory at Bhopal. Of these, 19% were found to be NiV positive by RT-PCR. Characteristics of NiV that increase the risk of it becoming a global pandemic include: humans are already susceptible; many NiV strains are capable of person-to-person transmission; and as an RNA virus, NiV has a high mutation rate. NiV has been found to survive for up to 4 days when subjected to various environmental conditions, including fruit bat urine and mango flesh. Whilst survival time was influenced by fluctuations in both temperature and pH, the ability for NiV to be spread by fomites could play a role in outbreak situations.
—————————–
Efficient Reverse Genetics Reveals Genetic Determinants of Budding and Fusogenic Differences between Nipah and Hendra Viruses and Enables Real-Time Monitoring of Viral Spread in Small Animal Models of Henipavirus Infection
02 January 2014
https://jvi.asm.org/content/89/2/1242
—————————–
COVID-19, Hendra and SARS: How scientists trace viruses through animals to their source
9 Feb 2021
https://www.abc.net.au/news/science/2021-02-10/covid-coronavirus-tracing-animal-origins-bats-sars-hendra/13081684
—————————–
Scientists Report Major Advance in the Treatment of Hendra Virus
2011
https://www.bumc.bu.edu/busm/2011/10/20/scientists-report-major-advance-in-the-treatment-of-hendra-virus/
—————————–
New strain of deadly Hendra virus discovered
9 Mar 2021
https://www.miragenews.com/new-strain-of-deadly-hendra-virus-525288/
—————————–
Hendra Virus to Coronavirus: WHO to Wake Up
May 7, 2020
https://dailytimes.com.pk/608386/hendra-virus-to-coronavirus-who-to-wake-up/
—————————–
Virulent Hendra virus has new symptoms in horses
23 July 2008
https://www.newscientist.com/article/mg19926663-500-virulent-hendra-virus-has-new-symptoms-in-horses/
—————————–
From Hendra to Wuhan: what has been learned in responding to emerging zoonotic viruses
February 11, 2020
https://www.thelancet.com/journals/lancet/article/PIIS0140-6736(20)30350-0/fulltext
—————————–
Nanotechnology provides novel solutions against zoonotic viruses
Aug 23 2021
https://www.news-medical.net/news/20210823/Nanotechnology-provides-novel-solutions-against-zoonotic-viruses.aspx
—————————–
Nigeria is already dealing with a deadlier viral outbreak than the coronavirus epidemic (Lassa Fever)
3-8-2020
https://qz.com/africa/1814567/coronavirus-less-deadly-than-nigeria-lassa-fever-viral-outbreak/
—————————————————————————-
COVID-19 is not a virus, but SARS-CoV-2 is
3-21-2020
https://virologydownunder.com/covid-19-is-not-a-virus-but-sars-cov-2-is/

For about two weeks we lived with, published using, and talked about, a disease-causing virus called the “novel coronavirus”. That name was always going to create problems like, what do we call the next one? After calling it the novel coronavirus, the World Health Organization (WHO) decided to refine the virus name. From around January 12th, they started calling it the 2019 novel coronavirus, 2019-nCoV. January 30th saw the WHO name the disease 2019-nCoV acute respiratory disease or 2019-nCoV-ARDS. But then in February, they changed that disease name to 2019 coronavirus disease or COVID-19. And the guys who are actually supposed to name viruses finally gave 2019-nCoV a new name; severe acute respiratory syndrome coronavirus 2: SARS-CoV-2 (It’s a shame HCoV-19 missed out[4]). As a result, understanding of the names is now a complete and utterly misunderstood mess at every level. Let’s dive into that and see if we can come up with some every day uses.
Confusing the matter
The WHO has chosen to not use the official virus name SARS-CoV-2 because…
From a risk communications perspective, using the name SARS can have unintended consequences in terms of creating unnecessary fear for some populations, especially in Asia which was worst affected by the SARS outbreak in 2003.
World Health Organization (WHO)
Okay. Sure. The WHO offer two examples of how they have decided to communicate with the public.
“the virus responsible for COVID-19” or “the COVID-19 virus”
World Health Organization (WHO)
Again, okay. The first one is a perfectly good workaround. The second though, while it may – to the author – imply we are talking about the virus that causes COVID-19, it really is just too economical with words.
A virus isn’t a disease, a disease isn’t a virus
An infectious disease is one you can catch from someone or somewhere else. But disease isn’t just one “thing” (I am not going to discuss “syndrome” here)
The word “disease” can be defined as a state of non-normal health characterised by two or more of the following criteria: recognized causal agent, identifiable group of signs and symptoms, consistent anatomic alterations.
Disease is a bucket term into which we pour the specific signs and symptoms, usually accompanied by how we test for the cause. We can add descriptions of the clinical course and outcomes. What disease is not though, is a virus. A virus – in the case of the disease called COVID-19 – is the cause of the signs and symptoms.
A virus is a distinct and transmissible agent that replicates and leads to all those things happening, in some direct or indirect fashion. A virus can be specifically tested for. How it causes disease can be examined and described.
Measures of the disease – cell counts, breathing rate, presence of pneumonia – can also be captured. But none of these is the virus.
When we test a sample of mucous or urine or stool or blood, we are looking for the virus or the effect of the virus. We are not looking for the disease. The disease is described by a doctor examining the patient also considering they test results. The Doctor then makes a diagnosis on the basis of those examinations and their judgment.
Some examples of confused understanding
Below are a few snippets from around the web. These weren’t hard to find but it’s worth noting that things have improved.
How can we use the words?
Here are a few example uses of the two labels, which may give you a feel for the difference.
The virus
I just got tested for SARS-CoV-2
That person is infected with SARS-CoV-2
SARS-CoV-2 was detected in their throat swab
They tested positive for SARS-CoV-2
The novel coronavirus (SARS-CoV-2) was in the community
If you spell out SARS-CoV-2 earlier
SARS-CoV-2 attaches to the ACE2 molecule
SARS-CoV-2 is an RNA virus with a lipid membrane
The disease
I was just diagnosed with COVID-19
A dry cough is a sign of COVID-19
COVID-19 was widespread in the community
COVID-19 can be severe in children and in adults
The virus causing COVID-19 can survive on surfaces for 48-72 hours
the new disease (COVID-19) was named by the WHO and caused by SARS-CoV-2
More clear now?
Hopefully, that makes a bit more sense. Feel free to send me some other examples in the comments below.
This isn’t the only example of different virus and disease names though. Some other examples include:
Human immunodeficiency virus (HIV) which causes AIDS
Herpes simplex virus (HSV) which causes cold sores
Varicella-zoster virus which causes chickenpox
Rhinoviruses which cause the common cold
There are also some more logical examples like:
Measles virus which causes measles
Dengue viruses which can cause dengue haemorrhagic fever
Influenza viruses which cause influenza
Since we’re all likely to have some time to think in the near future, why not use a bit of that time to get this straight?
—————————
The coronavirus isn’t alive. That’s why it’s so hard to kill.
3-23-2020
The science behind what makes this coronavirus so sneaky, deadly and difficult to defeat
https://www.washingtonpost.com/health/2020/03/23/coronavirus-isnt-alive-thats-why-its-so-hard-kill/
Viruses have spent billions of years perfecting the art of surviving without living — a frighteningly effective strategy that makes them a potent threat in today’s world.
That’s especially true of the deadly new coronavirus that has brought global society to a screeching halt. It’s little more than a packet of genetic material surrounded by a spiky protein shell one-thousandth the width of an eyelash, and it leads such a zombielike existence that it’s barely considered a living organism.
But as soon as it gets into a human airway, the virus hijacks our cells to create millions more versions of itself.
There is a certain evil genius to how this coronavirus pathogen works: It finds easy purchase in humans without them knowing. Before its first host even develops symptoms, it is already spreading its replicas everywhere, moving onto its next victim. It is powerfully deadly in some but mild enough in others to escape containment. And for now, we have no way of stopping it.
As researchers race to develop drugs and vaccines for the disease that has already sickened 350,000 and killed more than 15,000 people, and counting, this is a scientific portrait of what they are up against.
‘Between chemistry and biology’
Respiratory viruses tend to infect and replicate in two places: In the nose and throat, where they are highly contagious, or lower in the lungs, where they spread less easily but are much more deadly.
This new coronavirus, SARS-CoV-2, adeptly cuts the difference. It dwells in the upper respiratory tract, where it is easily sneezed or coughed onto its next victim. But in some patients, it can lodge itself deep within the lungs, where the disease can kill. That combination gives it the contagiousness of some colds, along with some of the lethality of its close molecular cousin SARS, which caused a 2002-2003 outbreak in Asia.
Another insidious characteristic of this virus: By giving up that bit of lethality, its symptoms emerge less readily than those of SARS, which means people often pass it to others before they even know they have it.
Viruses much like this one have been responsible for many of the most destructive outbreaks of the past 100 years: the flus of 1918, 1957 and 1968; and SARS, MERS and Ebola. Like the coronavirus, all these diseases are zoonotic — they jumped from an animal population into humans. And all are caused by viruses that encode their genetic material in RNA.
That’s no coincidence, scientists say. The zombielike existence of RNA viruses makes them easy to catch and hard to kill.
Outside a host, viruses are dormant. They have none of the traditional trappings of life: metabolism, motion, the ability to reproduce.
And they can last this way for quite a long time. Recent laboratory research showed that, although SARS-CoV-2 typically degrades in minutes or a few hours outside a host, some particles can remain viable — potentially infectious — on cardboard for up to 24 hours and on plastic and stainless steel for up to three days. In 2014, a virus frozen in permafrost for 30,000 years that scientists retrieved was able to infect an amoeba after being revived in the lab.
When viruses encounter a host, they use proteins on their surfaces to unlock and invade its unsuspecting cells. Then they take control of those cells’ molecular machinery to produce and assemble the materials needed for more viruses.
“It’s switching between alive and not alive,” said Gary Whittaker, a Cornell University professor of virology. He described a virus as being somewhere “between chemistry and biology.”

(The new coronavirus is one-thousandth the width of an eyelash in size and, like other viruses, is so molecularly simple that scientists barely consider it a living organism).
Among RNA viruses, coronaviruses — named for the protein spikes that adorn them like points of a crown — are unique for their size and relative sophistication. They are three times bigger than the pathogens that cause dengue, West Nile and Zika, and are capable of producing extra proteins that bolster their success.
“Let’s say dengue has a tool belt with only one hammer,” said Vineet Menachery, a virologist at the University of Texas Medical Branch. This coronavirus has three different hammers, each for a different situation.
Among those tools is a proofreading protein, which allows coronaviruses to fix some errors that happen during the replication process. They can still mutate faster than bacteria but are less likely to produce offspring so riddled with detrimental mutations that they can’t survive.
Meanwhile, the ability to change helps the germ adapt to new environments, whether it’s a camel’s gut or the airway of a human unknowingly granting it entry with an inadvertent scratch of her nose.
Scientists believe that the SARS virus originated as a bat virus that reached humans via civet cats sold in animal markets. This current virus, which can also be traced to bats, is thought to have had an intermediate host, possibly an endangered scaly anteater called a pangolin.
“I think nature has been telling us over the course of 20 years that, ‘Hey, coronaviruses that start out in bats can cause pandemics in humans, and we have to think of them as being like influenza, as long-term threats,’” said Jeffery Taubenberger, virologist with the National Institute of Allergy and Infectious Diseases.
Funding for research on coronaviruses increased after the SARS outbreak, but in recent years that funding has dried up, Taubenberger said. Such viruses usually simply cause colds and were not considered as important as other viral pathogens, he said.
The search for weapons
Once inside a cell, a virus can make 10,000 copies of itself in a matter of hours. Within a few days, the infected person will carry hundreds of millions of viral particles in every teaspoon of his blood.
The onslaught triggers an intense response from the host’s immune system: Defensive chemicals are released. The body’s temperature rises, causing fever. Armies of germ-eating white blood cells swarm the infected region. Often, this response is what makes a person feel sick.
Andrew Pekosz, a virologist at Johns Hopkins University, compared viruses to particularly destructive burglars: They break into your home, eat your food, use your furniture and have 10,000 babies. “And then they leave the place trashed,” he said.
Unfortunately, humans have few defenses against these burglars.
Most antimicrobials work by interfering with the functions of the germs they target. For example, penicillin blocks a molecule used by bacteria to build their cell walls. The drug works against thousands of kinds of bacteria, but because human cells don’t use that protein, we can ingest it without being harmed.
But viruses function through us. With no cellular machinery of their own, they become intertwined with ours. Their proteins are our proteins. Their weaknesses are our weaknesses. Most drugs that might hurt them would hurt us, too.
For this reason, antiviral drugs must be extremely targeted and specific, said Stanford virologist Karla Kirkegaard. They tend to target proteins produced by the virus (using our cellular machinery) as part of its replication process. These proteins are unique to their viruses. This means the drugs that fight one disease generally don’t work across multiple ones.
And because viruses evolve so quickly, the few treatments scientists do manage to develop don’t always work for long. This is why scientists must constantly develop new drugs to treat HIV, and why patients take a “cocktail” of antivirals that viruses must mutate multiple times to resist.
“Modern medicine is constantly needing to catch up to new emerging viruses,” Kirkegaard said.

(SARS-CoV-2 emerges from the surface of cells cultured in a lab).
SARS-CoV-2 is particularly enigmatic. Though its behavior is different from that of its cousin SARS, there are no obvious differences in the viruses’ spiky protein “keys” that allow them to invade host cells.
Understanding these proteins could be critical to developing a vaccine, said Alessandro Sette, head of the center for infectious disease at the La Jolla Institute for Immunology. Previous research has shown that the spike proteins on SARS are what trigger the immune system’s protective response. In a paper published this month, Sette found the same is true of SARS-CoV-2.
This gives scientists reason for optimism, according to Sette. It affirms researchers’ hunch that the spike protein is a good target for vaccines. If people are inoculated with a version of that protein, it could teach their immune system to recognize the virus and allow them to respond to the invader more quickly.
“It also says the novel coronavirus is not that novel,” Sette said.
And if SARS-CoV-2 is not so different from its older cousin SARS, then the virus is probably not evolving very fast, giving scientists developing vaccines time to catch up.
In the meantime, Kirkegaard said, the best weapons we have against the coronavirus are public health measures, such as testing and social distancing, and our own immune systems.
Some virologists believe we have one other thing working in our favor: the virus itself.
For all its evil genius and efficient, lethal design, Kirkegaard said, “the virus doesn’t really want to kill us. It’s good for them, good for their population, if you’re walking around being perfectly healthy.”
Evolutionarily speaking, experts believe, the ultimate goal of viruses is to be contagious while also gentle on their hosts — less a destructive burglar and more a considerate house guest.
That’s because highly lethal viruses like SARS and Ebola tend to burn themselves out, leaving no one alive to spread them.
But a germ that’s merely annoying can perpetuate itself indefinitely. One 2014 study found that the virus causing oral herpes has been with the human lineage for 6 million years. “That’s a very successful virus,” Kirkegaard said.
Seen through this lens, the novel coronavirus that is killing thousands across the world is still early in its life. It replicates destructively, unaware that there’s a better way to survive.
But bit by bit, over time, its RNA will change. Until one day, not so far in the future, it will be just another one of the handful of common cold coronaviruses that circulate every year, giving us a cough or sniffle and nothing more.
—————————–
You can’t kill the coronavirus. That’s OK. (Debated)
March 25, 2020
https://mashable.com/article/coronavirus-kill-dead-or-alive
“It’s to our own advantage to know our enemies as well as possible.”
Some viruses look like moon landers.
Called phages, they hijack bacteria by landing on the hapless cells and injecting them with a ream of genetic material. Then, the phages use the commandeered cells to multiply.
Similar to the new coronavirus, these phages are excellent parasites. They can be aggressive, dogged, and seem to act with purpose. Yet, many microbiologists who know viruses best say it’s a stretch to call any virus truly alive. And so, they can’t be killed — only disarmed, like pulling the plug on an appliance.
But today, with a rapidly spreading viral pandemic that’s stirring serious unease in American emergency rooms, it doesn’t really matter if a virus meets biologists’ definitions of dead or alive. Whatever these entities are, they’re powerful…
—————————–
Are Viruses Alive? – with Carl Zimmer
Nov 25, 2021
https://www.youtube.com/watch?v=Tryg5UCp6fI
—————————–
Scientists discover largest bacteria-eating virus. It blurs line between living and nonliving.
2-14-2020
https://www.livescience.com/largest-bacteriophage-discovered.html
Huge bacteria-killing viruses lurk in ecosystems around the world from hot springs to freshwater lakes and rivers. Now, a group of researchers has discovered some of these so-called bacteriophages that are so large and so complex that they blur the line between living and nonliving, according to new findings.
Bacteriophages, or “phages” for short, are viruses that specifically infect bacteria. Phages and other viruses are not considered living organisms because they can’t carry out biological processes without the help and cellular machinery of another organism.
—————————
Move Over Bacteria, Make Way for Protists
MAY 23, 2021
In the world of microbes, organisms like viruses and bacteria get a lot of attention. But now, researchers are beginning to study other unicellular life forms, like archaea, and protists. Protists live in many environments that contain water and include organisms like algae; other protists dwell in soil. This group is a kind of catch-all assigned to any eukaryote that is not an animal, fungi, or plant. It includes slime molds, diatoms, and amoebas, and many others. Though a few have been studied in-depth, like malaria-causing Plasmodium, we know very little about most others and more are being discovered all the time.

Scientists are learning more about the dynamic relationship between protists that live in soil and plants. It seems that protists and plants have a symbiotic relationship like the one shared by plants and some bacteria, according to new findings in the journal Microbiome.
—————————
Viruses 101: Why the new coronavirus is so contagious and how we can fight it
3-17-2020
Learn why the new coronavirus is so contagious. Is a cure or vaccine for COVID-19 coming? And will summer make a difference?
https://www.uchealth.org/today/viruses-101-why-the-new-coronavirus-is-so-contagious-how-to-fight-it/
—————————-
Coronavirus Outbreak? No Worries, Nanotechnology Is There to Help!
2020-01-28
https://statnano.com/news/67483/Coronavirus-Outbreak-No-Worries-Nanotechnology-Is-There-to-Help!
The ongoing coronavirus outbreak in Wuhan, China, causing rising death tolls, is a wake-up call for global health, in so far as many researchers have turned their focus to this growing threat.
Coronaviruses are a group of viruses that attack the upper and lower respiratory tracts in humans and cause a range of illnesses from the common cold to more serious forms such as severe acute respiratory syndrome (SARS) and the Middle East respiratory syndrome (MERS) which are life-threatening. These viruses can be transmitted to humans by different species of animals, including camels, cats, and probably bats.
Virions of coronavirus are in the form of spheres with an average diameter of 125 nm. They have a viral envelope and a positive-sense single-stranded RNA genome. Virus particles of coronavirus have four types of structural proteins, namely spike (S), membrane (M), envelope (E), and nucleocapsid (N) proteins, among which the S protein has a crucial role in attaching the virus to its host’s cells and enabling it to enter the cells; thus, an effective way of fighting this virus could be targeting the S protein’s mechanism of action by developing special drugs and inhibiting compounds.
Given their high specific surface area and the possibility of being functionalized with a wide range of functional groups, nanomaterials such as gold nanoparticles and carbon quantum dots (CQDs) are standout choices for interacting with viruses and preventing their entry into cells.
Recently, a group of researchers from the University of Lille, France, and Ruhr-University Bochum, Germany, showed that the CQDs functionalized with boronic acid ligands interfered with the function of coronavirus’s S protein and significantly inhibited its entry into the host cells. Their studies demonstrated that the addition of these nanomaterials to the cell culture medium, before and during infection with coronavirus, considerably reduced the infection rate of the cells. Surprisingly, after one viral life cycle – which is 5.5 hours for coronavirus – a great inhibition activity was also observed at the viral replication step.
These CQDs with an average diameter of 10 nm and excellent solubility in water can be perfect candidates for winning the battle against coronavirus, because they easily enter the cell through endocytosis and interact with the virus’s protein, thereby preventing viral genome replication.
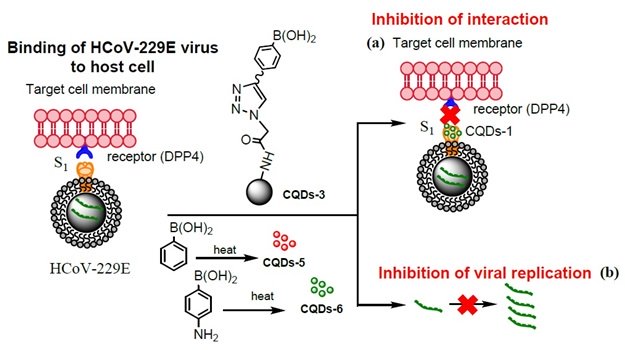
Influence of CQDs, prepared by hydrothermal carbonization, on the binding of HCoV- 229E virus to cells: (a) inhibition of protein S receptor interaction, (b) inhibition of viral RNA genome replication.
Other nanomaterials have also been found with the same antiviral effect. For instance, a Chinese research team has developed a novel class of peptide inhibitors based on gold nanorods, which selectively target coronavirus’s S protein and disrupt its activity.
Furthermore, the coronavirus vaccine has recently been prepared by means of nanotechnology; the researchers of Shizuoka University, Japan, succeeded in synthesizing virus-like particles (VLPs) in the form of nanovesicles using insect cells. These particles are very similar to coronavirus, except that they do not have the virus genome; once they enter into the host’s cells, they stimulate the cells’ immune system to fight the infection caused by this virus type.
In order to develop the coronavirus vaccine in the mentioned study, the team expressed the structural proteins of MERS coronavirus in silkworm larvae and Bm5 cells. The S protein of MERS coronavirus – without its transmembrane and cytoplasmic domains – was injected into the hemolymph of silkworm larvae and then purified via affinity chromatography. Afterwards, the purified proteins formed the nanoparticles that were able to attach to the receptor of coronavirus (called dipeptidyl peptidase 4) on the surface of the host cells. The E and M proteins were also expressed similarly, but an important point to consider was the fact that the S protein could only function provided that it was fully displayed on the VLPs; hence, the researchers used surfactant treatment and mechanical extrusion methods to develop S-protein-displaying nanovesicles with diameters of around 100 to 200 nm. Ultimately, the prepared VLP mimetic nanovesicles proved to be effective against MERS coronavirus, and indeed can be used as a nanoparticle-based vaccine to fight the ongoing coronavirus outbreak.
—————————
Nanotechnology and the fight against COVID-19
Jun 29 2021
https://www.news-medical.net/news/20210629/Nanotechnology-and-the-fight-against-COVID-19.aspx
—————————
Using the power of nanogold against cancer – and other cancer research at Georgia Tech
April 3, 2010
https://www.nanowerk.com/news/newsid=15631.php
—————————
Gold Nanoprisms as Optical Coherence Tomography Contrast Agents in the Second Near Infrared Window for Enhanced Angiography in Live Animals
May 16, 2018
https://www.biorxiv.org/content/10.1101/322545v1
—————————
Multi-functional stimuli sensitive polymer-gold nanocage carrier and preparation method thereof
2014
https://patents.google.com/patent/CN104258391A/en
Abstract
The invention belongs to the field of high polymer chemistry and biomedical engineering, and particularly relates to a multi-functional stimuli sensitive polymer-gold nanocage carrier and a preparation method thereof. The multi-functional carrier has a nucleus-shell-crown structure formed by a nucleus, a shell and a crown; the nucleus is formed by polymeric micelles, the shell is formed by gold nanocages, and the crown is formed by methoxy polyethylene glycol-branched polyethylenimine-lipoic acid; and the nano-carrier has a good photothermal conversion effect on the condition of 808 nm. The carrier has low toxicity for cells and can bear not only drugs but also genes or nucleic acids, thereby achieving the integration of carrier positioning, chemotherapy, thermal therapy and gene therapy and having a wide application value.
—————————
Targeted theranostics of lung cancer: PD-L1-guided delivery of gold nanoprisms with chlorin e6 for enhanced imaging and photothermal/photodynamic therapy
2020 Sep 29
https://pubmed.ncbi.nlm.nih.gov/33007481/
—————————
Spiral Hydroporator To Deliver Nanotechnologies Into Cells
March 18th, 2020
https://www.medgadget.com/2020/03/spiral-hydroporator-to-deliver-nanotechnologies-into-cells.html
—————————
Fabrication of RNA 3D Nanoprisms for Loading and Protection of Small RNAs and Model Drugs
2016
http://rnanano.osu.edu/Guo/Papers/2016%20-%20Advanced%20Materials%20-%20Khisamutdinov%20et%20al.%20-%20Fabrication%20of%20RNA%203D%20Nanoprisms%20for%20Loading%20and%20Protection%20of%20Small%20RNAs%20and%20Model%20D
—————————
Method of producing gold nanoprisms
2006
https://patents.google.com/patent/US7588624B2/en
—————————
Gold “nanoprisms” open new window into vessels and single cells
December 13, 2018
https://scopeblog.stanford.edu/2018/12/13/gold-nanoprisms-open-new-window-into-vessels-and-single-cells/
—————————
Polyvinylpyrrolidone-induced anisotropic growth of gold nanoprisms in plasmon-driven synthesis
2016
https://www.osti.gov/biblio/1290403-polyvinylpyrrolidone-induced-anisotropic-growth-gold-nanoprisms-plasmon-driven-synthesis
—————————
Tunable electronic lens and prisms using inhomogeneous nano scale liquid crystal droplets scale liquid crystal droplets
5-9-2006
https://stars.library.ucf.edu/cgi/viewcontent.cgi?referer=&httpsredir=1&article=1620&context=patents
—————————
An Efficient Near-Infrared Photothermal Therapy Agent by Using Ag @ Oxides Nanoprisms in for Cancer Therapy
April 2014
https://iovs.arvojournals.org/article.aspx?articleid=2270161
—————————
Near (NIR) and Far (FIR) Infrared Light, Photobiomodulation (PBM)
2020
https://www.advancedhealing.com/near-nir-and-far-fir-infrared-light-photobiomodulation-pbm/
—————————
Plasmon resonance in nanopatch antennas with triangular nanoprisms
2019
https://www.sciencedirect.com/science/article/abs/pii/S0030402619304206
—————————
Embedded plasmonic nanoprisms in polymer solar cells: Band-edge resonance for photocurrent enhancement
2020
https://aip.scitation.org/doi/10.1063/5.0002501
—————————
Enhancement of GdVO4:Eu3+ red fluorescence through plasmonic effect of silver nanoprisms on Si solar cell surface
May 21, 2019
Abstract
A comprehensive study on deposition and conjugation of red emitting phosphor (GdVO4 :Eu3+) and silver nanoprisms (Ag NP) on commercial single crystal silicon solar cell surface has been done to establish the optimum dielectric separating layer (PVA) sequence between consecutive
species through confocal fluorescence mapping and spectroscopy. Results show that up to 310% fluorescence enhancement could be achieved in optimal arrangement of emitter, PVA and Ag NP layers on Si cell surface. The current work shows the potential of plasmon enhanced down shifting fluorescence of rare-earth doped vanadate in enhancing performance of SiPV devices.
https://jart.icat.unam.mx/index.php/jart/article/view/640
—————————
Nanoparticle Material Melts Away to Reveal Drug Cargo
February 26th, 2018
At the Technion-Israel Institute of Technology, researchers have developed a new set of materials designed to deliver drugs inside the body and melt away when illuminated with light.
The materials are made of a polymer seeded with nano-sized gold shell nanoparticles. When light from a near-infrared (NIR) laser is applied to the material, it melts and reveals whatever it is ferrying.
One of the advantages of the material is that unlike many similar approaches, it relies on light frequencies that can penetrate fairly deep into the body without causing it damage compared to light of shorter wavelengths. Nevertheless, the energy delivered is enough to create enough heat to melt away the polymer, itself an FDA cleared material.
Each of the materials can have different melting points, allowing them to be produced specifically for certain applications. So far, though, this is still a laboratory project that will still involve creating actual melt-away particles that carry drugs before trying it on animal models.
Yet, the technology looks promising and may play a role in killing tumors, treating injuries, and acting as a foundation for new tissue to grow.



—————————
Cooperation of Hot Holes and Surface Adsorbates in Plasmon-Driven Anisotropic Growth of Gold Nanostars
2020
https://tsapps.nist.gov/publication/get_pdf.cfm?pub_id=930130
—————————
Plasmon-induced hot carrier science and technology
06 January 2015
https://www.nature.com/articles/nnano.2014.311
Abstract
The discovery of the photoelectric effect by Heinrich Hertz in 1887 set the foundation for over 125 years of hot carrier science and technology. In the early 1900s it played a critical role in the development of quantum mechanics, but even today the unique properties of these energetic, hot carriers offer new and exciting opportunities for fundamental research and applications. Measurement of the kinetic energy and momentum of photoejected hot electrons can provide valuable information on the electronic structure of materials. The heat generated by hot carriers can be harvested to drive a wide range of physical and chemical processes. Their kinetic energy can be used to harvest solar energy or create sensitive photodetectors and spectrometers. Photoejected charges can also be used to electrically dope two-dimensional materials. Plasmon excitations in metallic nanostructures can be engineered to enhance and provide valuable control over the emission of hot carriers. This Review discusses recent advances in the understanding and application of plasmon-induced hot carrier generation and highlights some of the exciting new directions for the field.
—————————
Heat transfer from nanoparticles: A corresponding state analysis
September 8, 2009
https://www.pnas.org/content/106/36/15113
—————————
Green synthesis of photomediated silver nanoprisms via a light-induced transformation reaction and silver nanoprism-impregnated bacteria cellulose films for use as antibacterial wound dressings
2019
Highlights
The photomediated silver nanoprisms (PAgNPrs) was synthesized in various shapes via excitation with 530 ± 20 nm LED lights.
The antibacterial effect of the PAgNPrs/BC-composite films against both gram-negative and gram-positive bacteria.
Non-toxicity of the PAgNPrs/BC composited films were confirmed by cytotoxicity test using human dermal fibroblasts.
PAgNPrs/BC composited films could be promising for use in wound dressing applications.
Abstract
Silver nanoprisms are used as antimicrobial agents for medical applications due to their large surface area, which provides better contact with microorganisms. However, silver nanoprisms are toxic to mammalian cells. To alleviate this problem, synthesis of silver nanoprisms via a light-induced transformation reaction with LED light was proposed. This reaction was ecofriendly and excluded hazards from the use of harmful oxidizing agents. Different shape and size of the as-grown silver nanocrystals could be distinguished by their colors. Results showed that the nano-octahedron photomediated silver nanoprisms showed the strongest bacterial inhibition with MIC and MBC value of 6.74, 6.74, 13.38, and 6.74 μg/mL against Escherichia coli, Pseudomonas aeruginosa, Enterococcus faecalis, and methicillin-resistant Staphylococcus aureus (MRSA), respectively. Silver nanoprisms were later impregnated within bacterial celluloses (BC) and the resulting composite films (PAgNPrs/BC) also exhibited the antibacterial activity against the four above microbes. Indirect cytotoxicity evaluation of the PAgNPrs/BC-composite films with human dermal fibroblasts proved that these films were non-toxic. All the results suggested that the PAgNPrs/BC-composite films could be used to care for certain infectious wounds.
https://www.sciencedirect.com/science/article/abs/pii/S1773224719309736
—————————
An electrically charged thin film patch used to promote wound healing
January 28, 2022
https://medicalxpress.com/news/2022-01-electrically-thin-patch-wound.html
—————————
Nanoporous Ag-Au Bimetallic Triangular Nanoprisms Synthesized by Galvanic Replacement for Plasmonic Applications
2018
https://www.hindawi.com/journals/jnm/2018/1263942/
—————————
Synthesis of Ag Nanoprisms with Precisely-tuned Localized Surface Plasmon Wavelengths by Sequential Irradiation of Light of Two Different Wavelengths
2020
https://www.journal.csj.jp/doi/abs/10.1246/cl.190888?journalCode=cl
—————————
Comparison of Triangular Silver Nanoprisms with Different Capping Agents and Structural Size for H2O2 Etching-Based Biosensors
2018
https://www.semanticscholar.org/paper/Comparison-of-Triangular-Silver-Nanoprisms-with-and-Huang-Yuan/23be43244fa0000084b5cdbae68c6cb764249bb9
—————————
Silver nanoprisms self-assembly on differently functionalized silica surface
2015
https://iopscience.iop.org/article/10.1088/1757-899X/77/1/012006/pdf
—————————
Time-Dependent Surface Plasmon Resonance Spectroscopy of Silver Nanoprisms in the Presence of Halide Ions
31 May 2010
https://chemistry-europe.onlinelibrary.wiley.com/doi/10.1002/cphc.200900842
—————————
Gold-reinforced silver nanoprisms on optical fiber tapers—A new base for high precision sensing
2016
https://aip.scitation.org/doi/full/10.1063/1.4953671
—————————
Solid-phase colorimetric sensing probe for bromide based on a tough hydrogel embedded with silver nanoprisms.
2020
https://www.sigmaaldrich.com/US/en/tech-docs/paper/1418401
—————————
Combined Use of Anisotropic Silver Nanoprisms with Different Aspect Ratios for Multi-Mode Plasmon-Exciton Coupling
16 January 2020
https://nanoscalereslett.springeropen.com/articles/10.1186/s11671-020-3248-8
—————————
Salicylazine activated plasmonic Silver Nanoprisms for deciphering Fe(II) & Fe(III) from their aqueous solutions
2021
https://pubs.rsc.org/en/content/articlelanding/2021/nj/d1nj03337a
—————————
Rapid thermal synthesis of silver nanoprisms with chemically tailorable thickness
2005
https://www.scholars.northwestern.edu/en/publications/rapid-thermal-synthesis-of-silver-nanoprisms-with-chemically-tail
—————————
Observation of Coalescence Process of Silver Nanospheres During Shape Transformation to Nanoprisms
2010
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3211880/
—————————
Habanero peppers provide ecofriendly way to synthesize silver nanoparticles
04.09.2020
https://www.innovations-report.com/life-sciences/a-spicy-silver-lining/
—————————
Selective Aggregation of Silver Nanoprisms Induced by Monohydrogen Phosphate and its Application for Colorimetric Detection of Chromium (III) Ions
03 July 2021
Abstract
In this study, we established a simple colorimetric method for visual detection of chromium (III) ions using aggregated silver nanoprisms induced by monohydrogen phosphate. The detection strategy had two steps, selective aggregation of silver nanoprisms with monohydrogen phosphate at high temperature, and anti aggregation of silver nanoprisms in the presence of chromium (III) ions. In this strategy, significant spectral shift of the localized surface plasmon resonance (LSPR) absorption peak of silver nanoprisms could be visualized with the color changed from yellow to blue. The spectral peak shift of silver nanoprisms in the anti-aggregation process was linear with the concentrations of the added chromium (III) ions in the range of 20.0–800.0 nM. Thus, a colorimetric method for chromium (III) ions detection was established with the detection limit of 3.6 nM. The experimental results of real water samples indicated that the colorimetric sensor was potential in environmental monitoring.
Results and Discussion
Characterization of Silver Nanoprisms
With reference to the literatures [24, 25], the synthesized silver nanoprisms had the characteristics of uniform size and shape distribution. In the synthesis process, we used LED lights to control the LSPR absorption peak of the silver nanoprisms at 575 nm, obtaining the uniformly distributed size and shape. As shown in Fig. 1a, the silver nanoprisms had three characteristic peaks at 340 nm, 420 nm and 575 nm, respectively. The small and sharp absorption peak at 340 nm was the out-of-plane fourth-order LSPR peak of the silver nanosheet, while the absorption peaks at 420 nm and 575 nm were the fourth-order and dipole LSPR peaks in the plane of the silver nanosheet [26], the result was consistent with that of the reported literature [27]. Under sunlight, it could be observed that the colloid of the silver nanoprisms was obviously blue (the inset of Fig. 1a). As shown in the transmission electron microscope (TEM) images (Fig. 1b-e), the silver nanoprisms exhibited triangular flake particles with the side length of about 80–100 nm, and maintaining good dispersion. In addition, the HRTEM image showed clear lattice fringes with a lattice spacing of about 0.25 nm (Fig. 1d), indicating that the silver nanoprisms had excellent crystallinity. As shown in Fig. 1e, the electron diffraction pattern showed f.c.c. structure of the silver nanoprisms (box-shaped spots corresponding to a lattice spacing of 0.144 nm, and circled spot corresponding to a lattice spacing of 0.25 nm).

UV–Vis absorption spectrum and photo image of silver nanoprisms at room temperature (a). TEM and High-resolution TEM images of silver nanoprisms (b–d). Electron diffraction pattern of silver nanoprisms (e)
Aggregation of the Silver Nanoprisms with Monohydrogen Phosphate at High Temperature
The silver nanoprisms had a triangular sheet structure. Many literatures reported that the edges and corners of silver nanoprisms had high chemical activity, therefore, external forces including temperature, time, external light source irradiation and additional chemical substances could all cause changes in the morphology of silver nanoprisms [6]. These factors rounded the edges and corners leading to silver nanoplates eventually. Simultaneously, the reduction of the aspect ratio of the silver nanoprisms resulted in a significant blue shift of the LSPR absorption peaks [28, 29], and the colloidal color of the silver nanoprisms changed from blue to yellow. As shown in Fig. 2, after being heated at 90 °C for 5.0 min, the LSPR absorption peak of the silver nanoprisms was blue-shifted from the original 575 to 498 nm (Fig. 2a), and the color of the silver nanoprisms sol changed from blue (Fig. 2b1) to pink (Fig. 2b2) accordingly. The transmission electron microscope (TEM) images demonstrated that the shape of the silver nanoprisms changed from a triangle (Fig. 2c) to a disc shape, and the dispersion was uniform (Fig. 2d).

UV–Vis absorption spectrum (a) and photo image (b) of silver nanoprisms at different conditions: 1, silver nanoprisms at 25 °C; 2, silver nanoprisms at 90 °C; 3, silver nanoprisms with HPO42− at 25 °C; 4, silver nanoprisms with HPO42− at 90 °C. Electron microscopy image of silver nanoprisms reacted at 25 °C for 5.0 min (c); reacted at 90 °C for 5.0 min (d); reacted at 25 °C for 5.0 min in the presence of HPO42− (e); reacted at 90 °C for 5.0 min in the presence of with HPO42− (f)
Anti-Aggregation Effect of Chromium (III) Ions
Our further studies found that the addition of chromium (III) ions would inhibit the aggregation of the silver nanoprisms induced by HPO42− ions, thereby reducing the blue-shift of the LSPR absorption peak. As shown in Fig. 5, when chromium (III) ions were added, and the silver nanoprisms mixture was heated at 90 °C for 5.0 min, the blue shift of the LSPR absorption peak was weakened (Fig. 5a). The transmission electron microscope image showed disc-shaped particles with a reduced degree of aggregation (Fig. 5e), and the color of the sol changing from yellow to pink (Fig. 5b3). As the concentration of added chromium (III) ions increased, the blue shift of the LSPR absorption peak was further weakened (as shown in Fig. 5a), and the transmission electron microscope image (TEM) showed disc-shaped particles and changing from aggregation to dispersion (as shown in Fig. 5f), the color of the sol was further deepened (Fig. 5b4) from pink.

UV–Vis absorption spectrum (a) and photo image (b) of silver nanoprisms at different conditions: 1, silver nanoprisms at 90 °C; 2, silver nanoprisms with HPO42− at 90 °C; 3, with HPO42− and Cr (III) ions (200 nM) at 90 °C; 4, with HPO42− and Cr (III) ions (600 nM) at 90 °C. Electron microscopy image of silver nanoprisms reacted at 90 °C for 5.0 min (c); reacted at 90 °C for 5.0 min in the presence of HPO42− (d); reacted at 90 °C for 5.0 min in the presence of HPO42− and 200 nM Cr (III) ions (e); reacted at 90 °C for 5.0 min in the presence of with HPO42− and 600 nM Cr (III) ions (f)
Analytical Parameter of Chromium(III) Ions
Since the anti-aggregation effect of chromium (III) ions on the silver nanoprisms could cause the blue shift of the LSPR absorption peak weakened, a quantitative method for detecting chromium (III) ions with silver nanoprisms as probes was established. As shown in Fig. 6, metal ions including Zn (II), Mg (II), Co (II), Cu (II), Fe (III), Hg (II), Cd (II), and Pb (II) etc. were tested under the optimal conditions. The results showed that this method had high selectivity. For typical cations, when the concentration was 50 times of those of chromium (III) ions, no obvious blue shift of the LSPR absorption peak was observed.
As shown in Fig. 7a, b, the degree of LSPR absorption peak shift weakening had a linear relationship with the concentrations of added chromium (III) ions. The calibration curve was λmax = 0.1229cCr (III) + 439.7453 (R2 = 0.9955) in the chromium (III) ions concentration range from 20.0 to 800.0 nM, with the limit of detection (LOD, 3σ) of 3.6 nM. It can be seen from Table 1, the concentration range and LOD were comparable to or better than most reported methods [31,32,33,34,35].

The absorption spectrum peak shifts (normalized) of silver nanoprisms with different concentrations of Cr (III) ions (a) and linear relationship between maximum absorption wavelength and concentration of Cr (III) ions (b). The color change of silver nanoprisms with different concentrations of Cr (III) ions (c). From 1 to 9, the concentration of Cr (III) ions is 60.0, 100.0, 200.0, 300.0, 400.0, 500.0, 600.0, 700.0 and 800.0 nM, respectively
Photoinduced conversion of silver nanospheres to nanoprisms
Nov 30 2001
https://www.scholars.northwestern.edu/en/publications/photoinduced-conversion-of-silver-nanospheres-to-nanoprisms
—————————
Silver Nanoprisms vs. Silver Nanospheres: The Battle Ensues.
July 15, 2011
http://ccsummerresearch.blogs.wm.edu/2011/07/15/silver-nanoprisms-vs-silver-nanospheres-the-battle-ensues/
—————————
Silver nanoparticle
https://en.wikipedia.org/wiki/Silver_nanoparticle
—————————
Differences between colloidal silver and ionic silver solutions
Nov 6 2019
How do silver nanoparticles disrupt cell walls to counter bacteria?
This is still a hotly debated subject and more research is required, but some research suggests that it is silver Ag+ ions that have a disruptive effect on the cellular membrane leading to death of the organism.

The problem here lies in ion delivery, as ingested solutions of ionic silver become silver compounds within 7 seconds of ingestion. Silver nanoparticles can travel through the human organism whilst releasing silver ions from their surface.
This process of oxidisation is slower than the direct ionic contact method, but in the cases where free ions such as chloride may be present (blood serum etc), silver nanoparticles are an effective delivery mechanism for silver ions due to their low reactivity potential. Whether antimicrobial property derives from the actual particle or their ion releasing capacity, the result is the same.
Nanosilver: Naughty or nice?
August 28, 2015
Questions abound about the possible effects of those tiny antibiotic particles that are showing up all over
https://www.sciencenewsforstudents.org/article/nanosilver-naughty-or-nice
—————————
Synthesis of uniform hexagonal Ag nanoprisms with controlled thickness and tunable surface plasmon bands
10 June 2019
https://link.springer.com/article/10.1007/s12613-019-1785-x
—————————
Metal chalcogenide hollow polar bipyramid prisms as efficient sulfur hosts for Na-S batteries
2020
https://theory.cm.utexas.edu/henkelman/pubs/aslam20_5242.pdf
—————————
Hierarchical Hollow Nanoprisms Based on Ultrathin Ni-Fe Layered Double Hydroxide Nanosheets with Enhanced Electrocatalytic Activity towards Oxygen Evolution
27 November 2017
https://onlinelibrary.wiley.com/doi/abs/10.1002/anie.201710877
—————————
Improved Efficiency of Organic Photovoltaic Cells by Incorporation of AuAg-Alloyed Nanoprisms
2017
https://ieeexplore.ieee.org/document/7896586
—————————
Multimodal cell tracking from systemic administration to tumour growth by combining gold nanorods and reporter genes
Jun 27, 2018
https://elifesciences.org/articles/33140
—————————
Second harmonic generation from phase-engineered metasurfaces of nanoprisms
2020
https://www.preprints.org/manuscript/202008.0304/v1/download
—————————
Synthesis, Characterization, and Self-Assembly of Gold Nanorods and Nanoprisms
2010
https://digitalcommons.usf.edu/etd/3446/
—————————
Polymorphic Assembly from Beveled Gold Triangular Nanoprisms
2017
https://chenlab.matse.illinois.edu/files/2016/07/Kim-2017-Polymorphic-Assembly-from-Beveled-Gold-Triangular-Nanoprisms.pdf
—————————
DNA-induced assembly of gold nanoprisms and polystyrene beads into 3D plasmonic SERS substrates
2021
https://iopscience.iop.org/article/10.1088/1361-6528/abbc22
—————————
Tailoring the synthesis and heating ability of gold nanoprisms for bioapplications
2012 Feb 9
https://pubmed.ncbi.nlm.nih.gov/22260484/
—————————
SERS detection of explosive agent by macrocyclic compound functionalized triangular gold nanoprisms
April 2011
https://analyticalsciencejournals.onlinelibrary.wiley.com/doi/10.1002/jrs.2932
—————————
Biological Synthesis of Colloidal Gold Nanoprisms Using Penicillium citrinum MTCC9999
2013
https://www.scirp.org/html/3-3200263_29846.htm
—————————
The effect of iodide on the synthesis of gold nanoprisms
26 Jan 2015.
https://www.tandfonline.com/doi/pdf/10.1080/17458080.2014.1003340
—————————
A gold–silicon potential fitted to the binary phase diagram
15 January 2010
https://iopscience.iop.org/article/10.1088/0953-8984/22/5/055401
—————————
Gold Nanoparticles Can Remote Control the Brain
March 27, 2015
It’s just the latest twist in nanotech that is using gold as medicine
https://www.smithsonianmag.com/smart-news/heated-gold-nanoparticles-let-scientists-remote-control-brain-180954789/
—————————
Nanoparticles: Building Blocks for Nanotechnology
2004
https://books.google.co.in/books/about/Nanoparticles.html?id=Aw4bbm8TcXkC
—————————
Nanomedicine
https://en.wikipedia.org/wiki/Nanomedicine
Nanomedicine is the medical application of nanotechnology. Nanomedicine ranges from the medical applications of nanomaterials and biological devices, to nanoelectronic biosensors, and even possible future applications of molecular nanotechnology such as biological machines. Current problems for nanomedicine involve understanding the issues related to toxicity and environmental impact of nanoscale materials (materials whose structure is on the scale of nanometers, i.e. billionths of a meter).
—————————
Nanoparticle
https://en.wikipedia.org/wiki/Nanoparticle
—————————
Nanofabrication using thermomechanical nanomolding
November 29, 2021
https://phys.org/news/2021-11-nanofabrication-thermomechanical-nanomolding.html
—————————
Nanomaterials
What are the potential health effects of nanomaterials?
2009
https://ec.europa.eu/health/scientific_committees/opinions_layman/nanomaterials/en/l-2/4.htm
—————————
Nanotechnologies
What are potential harmful effects of nanoparticles?
30-7-2021
https://copublications.greenfacts.org/en/nanotechnologies/l-2/6-ealth-effects-nanoparticles.htm
—————————
Toxicity and Environmental Risks of Nanomaterials: Challenges and Future Needs
2009
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2844666/
—————————
Heavy Metal Toxicity Increases Your Risk of Electromagnetic Sensitivity
May 28, 2008
https://www.wellnesscenter.net/resources/Heavy_Metal_Toxicity.htm
—————————
Nanochips and smart dust – dangerous new face of human microchipping agenda
October 20, 2017
Nanochips and Smart Dust are the new technological means for the advancement of the human microchipping agenda. Due to their incredibly tiny size, both nanochips and Smart dust have the capacity to infiltrate the human body, become lodged within, and begin to set up a synthetic network on the inside which can be remotely controlled from the outside. Needless to say, this has grave freedom, privacy and health implications, because it means the New World Order would be moving from controlling the outside world (environment/society) to controlling the inside world (your body). This article explores what the advent of nanochips and Smart dust could mean for you.
https://nexusnewsfeed.com/article/human-rights/nanochips-and-smart-dust-dangerous-new-face-of-human-microchipping-agenda/
—————————
Mesoporous silica nanoparticles in medicine—Recent advances
2012
https://www.sciencedirect.com/science/article/abs/pii/S0169409X12002487
—————————
Silica nanoparticles trigger the vascular endothelial dysfunction and prethrombotic state via miR-451 directly regulating the IL6R signaling pathway
11 April 2019
https://particleandfibretoxicology.biomedcentral.com/articles/10.1186/s12989-019-0300-x
—————————
Structural and Chemical Modifications Towards High-Performance of Triboelectric Nanogenerators
30 July 2021
https://nanoscalereslett.springeropen.com/articles/10.1186/s11671-021-03578-z
—————————
Hybridized Nanogenerators for Multifunctional Self-Powered Sensing: Principles, Prototypes, and Perspectives
December 18, 2020
https://www.cell.com/iscience/fulltext/S2589-0042(20)31010-5
—————————
Nano-and Micromotors Designed for Cancer Therapy
September 2019
https://www.researchgate.net/publication/335944007_Nano-and_Micromotors_Designed_for_Cancer_Therapy
—————————
Drug delivery and nanoparticles: Applications and hazards
2008
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2527668/
—————————
Shape-morphing microrobots deliver drugs to cancer cells
November 17, 2021
https://www.sciencedaily.com/releases/2021/11/211117103858.htm
—————————
Tiny Robots Can Clear Clogged Arteries
July 1, 2015
Engineers at Drexel University are developing micro-swimmers that loosen arterial plaque and release drugs into the bloodstream to prevent future buildup
https://www.smithsonianmag.com/innovation/tiny-robots-can-clear-clogged-arteries-180955774/
—————————
Here’s how nanoparticles could help us get closer to a treatment for COVID-19
March 4, 2020
https://news.northeastern.edu/2020/03/04/heres-how-nanoparticles-could-help-us-get-closer-to-a-treatment-for-covid-19/
There is no vaccine or specific treatment for COVID-19, the disease caused by the severe acute respiratory syndrome coronavirus 2, or SARS-CoV-2.
Since the outbreak began in late 2019, researchers have been racing to learn more about SARS-CoV-2, which is a strain from a family of viruses known as coronavirus for their crown-like shape.
Northeastern chemical engineering professor Thomas Webster, who specializes in developing nano-scale medicine and technology to treat diseases, is part of a contingency of scientists that are contributing ideas and technology to the Centers for Disease Control and Prevention to fight the COVID-19 outbreak.
The idea of using nanoparticles, Webster says, is that the virus behind COVID-19 consists of a structure of a similar scale as his nanoparticles. At that scale, matter is ultra-small, about ten thousand times smaller than the width of a single strand of hair.
Webster is proposing particles of similar sizes that could attach to SARS-CoV-2 viruses, disrupting their structure with a combination of infrared light treatment. That structural change would then halt the ability of the virus to survive and reproduce in the body.
“You have to think in this size range,” says Webster, who is Art Zafiropoulo Chair in Engineering at Northeastern. “In the nanoscale size range, if you want to detect viruses, if you want to deactivate them.”
Finding and neutralizing viruses with nanomedicine is at the core of what Webster and other researchers call theranostics, which focuses on combining therapy and diagnosis. Using that approach, his lab has specialized in nanoparticles to fight the microbes that cause influenza and tuberculosis.
“It’s not just having one approach to detect whether you have a virus and another approach to use it as a therapy,” he says, “but having the same particle, the same approach, for both your detection and therapy.”
SARS-CoV-2 spreads mostly through tiny droplets of viral particles—from breathing, talking, sneezing, coughing—that enter the body through the eyes, mouth, or nose. Preliminary research also suggests that those germs may survive for days when they attach themselves to countertops, handrails, and other hard surfaces.
That’s one reason to make theranostics with nanoparticles the focus of the COVID-19 outbreak, Webster says.
Nanoparticles can disable these pathogens even before they break into the body, as they hold on to different objects and surfaces. His lab has developed materials that can be sprayed on objects to form nanoparticles and attack viruses.
“Even if it was on a surface, on someone’s countertop, or an iPhone,” he says. “It doesn’t mean anything because it’s not the active form of that virus.”
That same technology can be fine-tuned and tweaked to target a wide range of viruses, bacteria, and other pathogens. Unlike other novel drugs with large molecular structures, nanoparticles are so small that they can move through our body without disrupting other functions, such as those of the immune system.
“Almost like a surveyor, they can go around your bloodstream,” Webster says. “They can survey your body much easier and under much longer times and try and detect viruses.”
To do all that, the CDC needs to know the specifics about what kind of structure is needed to neutralize SARS-CoV-2, Webster says. That information isn’t public yet.
“You have to identify what we need to put in our nanoparticle to attract it to that virus,” he says. “The CDC must know that, because they’ve developed a kit that can determine if you have [COVID-19], versus influenza, or something else.”
An alternative to nanomedicine is producing synthetic molecules. But Webster says that tactic presents some challenges. In the case of chemotherapies used to treat cancer cells, such synthetic drugs can cause severe side effects that kill cancer cells, as well as other cells in the body.
“The same thing could be happening with synthetic chemistry to treat a virus, where molecules are killing a lot more than just that virus,” Webster says.
Still, Webster acknowledges that there aren’t many researchers focusing on nanoparticles to kill viruses.
One of the main reasons for the lack of those solutions is that the same benefits that make nanoparticles ideal to fight infectious diseases also make them a concern for the U.S. Federal Drug Administration.
Because of their size, nanoparticles are pervasive (too pervasive, maybe) to seep through other parts of the body. To reduce that risk, Webster’s lab has focused on using iron oxide. Particles of that make up entail chemistry that is already natural to our bodies and diets.
“Even if you have a viral infection, you need more iron, because you could be anemic depending on how bad the infection is,” Webster says. “We’re actually developing these nanoparticles out of chemistries that can help your health.”
And, he says, iron-based nanoparticles could be directed with magnetic fields to target specific organs in the body, such as lungs and other areas susceptible to respiratory complications after contracting viral infections. That too, Webster says, is something that you couldn’t do with a novel synthetic molecule.
“Really, what this all means is that we just have to do the studies to show those iron nanoparticles are not going into the brain or the kidney,” Webster says, “that these nanoparticles are going exactly where you want them to go to the virus.”
————————
Coronavirus outbreak: doctors use robot to treat first known US patient
Jan 2020
https://www.theguardian.com/us-news/2020/jan/22/coronavirus-doctors-use-robot-to-treat-first-known-us-patient
———————————————
DNA Nanobots Used Bacteria’s Chemical Communication and This Enables Intelligent Nanobot Swarms
11-16-2019

https://www.nextbigfuture.com/2019/11/dna-nanobots-used-bacterias-chemical-communication-and-this-enables-intelligent-nanobot-swarms.html
—————————
New nanobots that ‘fly’ through the body could perform tiny surgeries
6 Nov 2019
https://www.siliconrepublic.com/machines/nanobots-birds-fly-human-body
—————————
‘Nanobot’ viruses tag and round up bacteria in food and water
March 27, 2018
Tweaking DNA and adding magnetic nanoparticles creates a new tool to test for contaminants
https://www.sciencenews.org/article/nanobot-viruses-tag-and-round-bacteria-food-and-water
—————————
Levitating and colliding liquid drops
January 14, 2022
https://phys.org/news/2022-01-levitating-colliding-liquid.html
—————————
Scientists capture a ‘quantum tug’ between neighboring water molecules
August 25, 2021
Summary:
Researchers have made the first direct observation of how hydrogen atoms in water molecules tug and push neighboring water molecules when they are excited with laser light.
https://www.sciencedaily.com/releases/2021/08/210825113614.htm
—————————
Genetically engineered virus ‘cures’ patients of skin cancer
2015
The new therapy has far fewer side effects because it is harnassing the body’s own immune system

(Genetically engineering the herpes virus so that it attacks skin cancer cells has potentially cured some patients, scientists have said).
https://www.telegraph.co.uk/news/science/science-news/11631626/v.html
—————————-
Anti-cancer virus fits tumor receptor like a ‘key in a lock’
2018
https://www.sciencedaily.com/releases/2018/10/181029150937.htm
—————————–
Oncolytic Virus Therapy: Using Tumor-Targeting Viruses to Treat Cancer
2018
https://www.cancer.gov/news-events/cancer-currents-blog/2018/oncolytic-viruses-to-treat-cancer
For more than a century, doctors have been interested in using viruses to treat cancer, and in recent years a small but growing number of patients have begun to benefit from this approach.
Some viruses tend to infect and kill tumor cells. Known as oncolytic viruses, this group includes viruses found in nature as well as viruses modified in the laboratory to reproduce efficiently in cancer cells without harming healthy cells.
To date, only one oncolytic virus—a genetically modified form of a herpesvirus for treating melanoma—has been approved by the Food and Drug Administration (FDA), though a number of viruses are being evaluated as potential treatments for cancer in clinical trials.

(An immunotherapy approach using the Maraba virus (above) and checkpoint inhibitors cured aggressive breast cancer in mice).
——————————-
Oncolytic targeting of androgen-sensitive prostate tumor by the respiratory syncytial virus (RSV): consequences of deficient interferon-dependent antiviral defense
28 January 2011
https://bmccancer.biomedcentral.com/articles/10.1186/1471-2407-11-43
——————————-
Imaging Oncolytic Virus Infection in Cancer Cells
2020
https://resources.perkinelmer.com/lab-solutions/resources/docs/APP_103134_OncolyticViruses.pdf
——————————-
Targeting the tumor stroma with an oncolytic adenovirus secreting a fibroblast activation protein-targeted bispecific T-cell engager
2019
https://jitc.bmj.com/content/7/1/19
——————————-
Tumor-Specific Targeting With Modified Sindbis Viral Vectors: Evaluation with Optical Imaging and Positron Emission Tomography In Vivo
31 July 2012
https://link.springer.com/article/10.1007/s11307-012-0585-8
——————————-
Oncolytic Maraba virus armed with tumor antigen boosts vaccine priming and reveals diverse therapeutic response patterns when combined with checkpoint blockade in ovarian cancer
17 July 2019
https://jitc.biomedcentral.com/articles/10.1186/s40425-019-0641-x
——————————-
Oncolytic targeting of androgen-sensitive prostate tumor by the respiratory syncytial virus (RSV): consequences of deficient interferon-dependent antiviral defense
28 January 2011
https://bmccancer.biomedcentral.com/articles/10.1186/1471-2407-11-43
——————————-
Imaging Oncolytic Virus Infection in Cancer Cells
2020
https://resources.perkinelmer.com/lab-solutions/resources/docs/APP_103134_OncolyticViruses.pdf
——————————-
Targeting the tumor stroma with an oncolytic adenovirus secreting a fibroblast activation protein-targeted bispecific T-cell engager
2019
https://jitc.bmj.com/content/7/1/19
——————————-
Tumor-Specific Targeting With Modified Sindbis Viral Vectors: Evaluation with Optical Imaging and Positron Emission Tomography In Vivo
31 July 2012
https://link.springer.com/article/10.1007/s11307-012-0585-8
——————————-
Oncolytic Maraba virus armed with tumor antigen boosts vaccine priming and reveals diverse therapeutic response patterns when combined with checkpoint blockade in ovarian cancer
17 July 2019
https://jitc.biomedcentral.com/articles/10.1186/s40425-019-0641-x
——————————-
The Use of a Tropism-Modified Measles Virus in Folate Receptor–Targeted Virotherapy of Ovarian Cancer
October 2006
https://clincancerres.aacrjournals.org/content/12/20/6170
——————————-
Targeting Visceral Fat by Intraperitoneal Delivery of Novel AAV Serotype Vector Restricting Off-Target Transduction in Liver
2017
https://www.sciencedirect.com/science/article/pii/S2329050117300773
——————————-
Sindbis virus – an effective targeted cancer therapeutic
2004
https://www.sciencedirect.com/science/article/abs/pii/S0167779904002240
——————————-
Synergistic anti-tumor effects between oncolytic vaccinia virus and paclitaxel are mediated by the IFN response and HMGB1
26 August 2010
https://www.nature.com/articles/gt2010121
——————————-
Oncolytic Virus-Mediated Targeting of PGE2 in the Tumor Alters the Immune Status and Sensitizes Established and Resistant Tumors to Immunotherapy
2016
https://www.sciencedirect.com/science/article/pii/S1535610816302173
——————————-
Evaluation of Tumor Specificity and Immunity of Thymidine Kinase-Deleted Vaccinia Virus Guang9 Strain
2020
https://pubmed.ncbi.nlm.nih.gov/32801778/
——————————-
Genetically engineering NK cells can destroy tumor-regenerating glioblastoma stem cells
Jun 17 2021
Preclinical research from The University of Texas MD Anderson Cancer Center finds that although glioblastoma stem cells (GSCs) can be targeted by natural killer (NK) cells, they are able to evade immune attack by releasing the TFG-β signaling protein, which blocks NK cell activity. Deleting the TFG-β receptor in NK cells, however, rendered them resistant to this immune suppression and enabled their anti-tumor activity.
The findings, published today in the Journal of Clinical Investigation, suggest that engineering NK cells to resist immune suppression may be a feasible path toward using NK cell-based immunotherapies for treating glioblastoma.
https://www.news-medical.net/news/20210617/Genetically-engineering-NK-cells-can-destroy-tumor-regenerating-glioblastoma-stem-cells.aspx
——————————-
Genetically modified virus may shrink incurable brain cancers
April 2019
https://www.newscientist.com/article/2200543-genetically-modified-virus-may-shrink-incurable-brain-cancers/
People with incurable melanomas and brain or breast cancers are to get injections of tumour-fighting viruses.
The trial will test the safety of a virus that has been engineered to shrink tumours – an approach that holds promise for a range of cancers, including deadly brain tumours.
The idea of using viruses to kill cancers goes back more than a century, inspired by anecdotal reports of some people with viral infections being cured of malignancies. But turning viruses that can infect and kill human cells into …

(Tiny phages that usually infect microbes could fight cancer).
——————————
Studying resistance to therapy in BRAF-mutated brain tumors
September 15, 2021
https://medicalxpress.com/news/2021-09-resistance-therapy-braf-mutated-brain-tumors.html
——————————
The Rapid Development and Early Success of Covid 19 Vaccines Have Raised Hopes for Accelerating the Cancer Treatment Mechanism
March 1, 2021
https://pubmed.ncbi.nlm.nih.gov/33818952/
——————————
New tumor cell tracking system aims to understand cancer treatment resistance
August 13, 2021
https://medicalxpress.com/news/2021-08-tumor-cell-tracking-aims-cancer.html
——————————
Potential therapeutic agents to COVID-19: An update review on antiviral therapy, immunotherapy, and cell therapy
Marcgh 16, 2021
https://pubmed.ncbi.nlm.nih.gov/33774315/
——————————
Development of COVID-19 vaccines utilizing gene therapy technology
2021
Abstract
There is currently an outbreak of respiratory disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Coronavirus disease 2019 (COVID-19) is caused by infection with SARS-CoV-2. Individuals with COVID-19 have symptoms that are usually asymptomatic or mild in most initial cases. However, in some cases, moderate and severe symptoms have been observed with pneumonia. Many companies are developing COVID-19 vaccine candidates using different technologies that are classified into four groups (intact target viruses, proteins, viral vectors and nucleic acids). For rapid development, RNA vaccines and adenovirus vector vaccines have been urgently approved, and their injection has already started across the world. These types of vaccine technologies have been developed over more than 20 years using translational research for use against cancer or diseases caused by genetic disorders but the COVID-19 vaccines are the first licensed drugs to prevent infectious diseases using RNA vaccine technology. Although these vaccines are highly effective in preventing COVID-19 for a short period, safety and efficiency evaluations should be continuously monitored over a long time period. As the time of writing, more than 10 projects are now in phase 3 to evaluate the prevention of infection in double-blind studies. Hopefully, several projects may be approved to ensure high-efficiency and safe vaccines.
https://pubmed.ncbi.nlm.nih.gov/33772572/
——————————
New gene-delivering workhorse could make gene therapy safer, more effective for muscle diseases
Sep 9 2021
https://www.news-medical.net/news/20210909/New-gene-delivering-workhorse-could-make-gene-therapy-safer-more-effective-for-muscle-diseases.aspx
——————————
CRISPR pioneer Feng Zhang’s latest work delivers mRNA, gene therapy with a human protein
Aug 19, 2021
https://www.fiercebiotech.com/research/crispr-pioneer-feng-zhang-s-latest-work-delivers-mrna-gene-therapy-human-protein
COVID-19 mRNA vaccines and existing gene therapies, including those built with the CRISPR-Cas9 gene-editing tool, are delivered into cells with viral vectors or lipid nanoparticles. A research team led by CRISPR pioneer Feng Zhang, Ph.D., of the Broad Institute has developed a new mRNA delivery system that harnesses a human protein.
The system, dubbed SEND, leverages the ability of a human protein called PEG10 to bind to its own mRNA and form a protective capsule around it. In a new study published in Science, Zhang and colleagues engineered PEG10 to take on RNA cargoes of their choice and successfully delivered the system to mouse and human cells.
The findings support SEND as an efficient delivery platform for RNA-based gene therapies that can be repeatedly dosed, the researchers suggested. Because SEND uses a protein that’s produced naturally in the body, it may not trigger immune responses that can render gene therapies ineffective, the team said.
PEG10 is a retroviruslike protein, expressed by a gene that integrated itself into the human genome through ancestral viral infection. A common feature of retroviruses and retroelements is a core structural gene known as “gag.” In mammals, equivalents of gag are known to form a protective shell to shield their genetic materials and transfer mRNA between cells.
This unique feature of these retroviruslike proteins inspired the idea that they could be effective delivery vehicles for gene therapies. In the current study, Zhang and colleagues searched human and mouse genomes and found 19 gag-derived genes that were present in both humans and mice. Further analysis pointed to PEG10 as an efficient delivery tool; the cells released significantly more PEG10 particles than other proteins, and the particles mostly contained their own mRNA.
To develop the delivery system, the researchers first identified the minimal sequences, or “signals,” on PEG10 mRNA’s untranslated region that were required to trigger efficient packaging and functional transfer of the mRNA. So the researchers knew what genetic elements to keep when reprogramming PEG10’s mRNA to carry genes of interest.
To improve the construct, the researchers added proteins, called “fusogens,” which facilitate the fusion of cells as a critical step for transferring genomic material. These cell surface proteins also allow researchers to direct PEG10 to a particular type of cell, tissue or organ.
The team tested SEND with the CRISPR-Cas9 system to edit specific genes. In CRISPR-Cas9, a short guide RNA sequence helps locate the target gene, and the Cas9 protein makes the cut.
Delivering a large Cas9 cargo into mouse cell lines that expressed a single guide RNA (sgRNA) led to about 60% insertions and deletions in recipient cells, the team reported. Co-packing the Cas9 with the sgRNA using the human SEND system, the scientists reported about 40% edits at a specific location on a chromosome.
Zhang is recognized as a trailblazer in the development of CRISPR and has mostly focused his work on improving the gene-editing tool. For example, he was a co-founder of Beam Therapeutics, which is working on CRISPR-based tools that enable the precise editing of specific base pairs of DNA.
Zhang and his researchers believe SEND could offer a new way to deliver RNAs as therapeutics or as a gene-editing tool to target problematic DNA. “By mixing and matching different components in the SEND system, we believe that it will provide a modular platform for developing therapeutics for different diseases,” Zhang said in a statement.
One problem with the currently used viral vector delivery platforms is that they only allow dosing once, because the immune system may recognize the vector and launch a strong attack on second administration. Given that SEND uses a human protein, it may not trigger an immune response and could therefore be repeatedly dosed, “which greatly expands the applications for nucleic acid therapy,” the researchers wrote in the study.
Next, the team will test SEND in animals and further improve the system to deliver RNA cargoes to different cells and tissues. They will also explore using other components in the human body to potentially add to the system.
“The realization that we can use PEG10, and most likely other proteins, to engineer a delivery pathway in the human body to package and deliver new RNA and other potential therapies is a really powerful concept,” Zhang said.
——————————
Mammalian retrovirus-like protein PEG10 packages its own mRNA and can be pseudotyped for mRNA delivery
19 Aug 2021
https://www.science.org/doi/10.1126/science.abg6155
——————————
CRISPR pioneer Zhang targets brain diseases with new RNA-editing system
Jul 11, 2019
https://www.fiercebiotech.com/research/crispr-pioneer-zhang-targets-brain-diseases-new-rna-editing-system
——————————
Apellis, Beam ink $75M deal to bring gene-editing meds to complement diseases
Jun 30, 2021
https://www.fiercebiotech.com/biotech/apellis-beam-ink-75m-deal-to-bring-gene-editing-meds-to-complement-diseases
——————————
Worse Than the Disease? Reviewing Some Possible Unintended Consequences of the mRNA Vaccines Against COVID-19
May 10, 2021
Operation Warp Speed brought to market in the United States two mRNA vaccines, produced by Pfizer and Moderna. Interim data suggested high efficacy for both of these vaccines, which helped legitimize Emergency Use Authorization (EUA) by the FDA. However, the exceptionally rapid movement of these vaccines through controlled trials and into mass deployment raises multiple safety concerns. In this review we first describe the technology underlying these vaccines in detail. We then review both components of and the intended biological response to these vaccines, including production of the spike protein itself, and their potential relationship to a wide range of both acute and long-term induced pathologies, such as blood disorders, neurodegenerative diseases and autoimmune diseases. Among these potential induced pathologies, we discuss the relevance of prion-protein-related amino acid sequences within the spike protein. We also present a brief review of studies supporting the potential for spike protein “shedding”, transmission of the protein from a vaccinated to an unvaccinated person, resulting in symptoms induced in the latter. We finish by addressing a common point of debate, namely, whether or not these vaccines could modify the DNA of those receiving the vaccination. While there are no studies demonstrating definitively that this is happening, we provide a plausible scenario, supported by previously established pathways for transformation and transport of genetic material, whereby injected mRNA could ultimately be incorporated into germ cell DNA for transgenerational transmission. We conclude with our recommendations regarding surveillance that will help to clarify the long-term effects of these experimental drugs and allow us to better assess the true risk/benefit ratio of these novel technologies.
https://ijvtpr.com/index.php/IJVTPR/article/view/23
——————————
Synthesized Nanoparticles Act As Artificial Viruses for Gene Therapy
04/09/2015
Researchers of the Nanobiology Unit from the Universitat Autònoma de Barcelona (UAB) Institute of Biotechnology and Biomedicine, led by Antonio Villaverde, managed to create artificial viruses, protein complexes with the ability of self-assembling and forming nanoparticles which are capable of surrounding DNA fragments, penetrating the cells and reaching the nucleus in a very efficient manner, where they then release the therapeutic DNA fragments. The achievement represents an alternative with no biological risk to the use of viruses in gene therapy.
http://www.cemag.us/news/2015/04/synthesized-nanoparticles-act-artificial-viruses-gene-therapy
——————————————
PROTACs: A New Type of Drug That Can Target All Disease-Causing Proteins
June 11, 2015
http://scitechdaily.com/protacs-a-new-type-of-drug-that-can-target-all-disease-causing-proteins/
A newly published study from Yale University details the discovery of a new type of drug, called Proteolysis Targeting Chimeras (PROTACs), which can target all disease-causing proteins.
Current drugs block the actions of only about a quarter of known disease-causing proteins, but Yale University researchers have developed a technology capable of not just inhibiting, but destroying every protein it targets.
The new type of drug, called Proteolysis Targeting Chimeras (PROTACs), also can continue to destroy mutant proteins in mouse tumors, according to a new study published June 10 in the journal Nature Chemical Biology.
“This new drug modality culminates a decade of work in the field by my lab,” said Craig Crews, the Lewis B. Cullman Professor of Molecular, Cellular, and Developmental Biology and senior author of the paper, which was done in collaboration with scientists from GlaxoSmithKline and Arvinas, LLC.
Almost all current drugs are small molecules designed to fit into the folds of disease-causing proteins and inhibit their function. High doses are often needed to ensure that protein function is blocked sufficiently to produce therapeutic results, which in turn can produce harmful side effects.
In contrast, PROTACs engage the cells’ own protein degradation machinery to destroy targeted proteins by tagging them for removal and can do so multiple times, meaning it can work at lower doses, the authors say. This suggests this new type of drug has not only the potential to target proteins that are not currently “pharmaceutically vulnerable” but could do so safely, Crews said.
“This is a game-changer for drug development,” Crews said.
—————————————-
Nanorobot takes on hepatitis C virus, wins
2012
https://newatlas.com/nanoparticles-hepatitis-c-university-florida/23379/
————————-
Nanorobot Hardware Architecture for Medical Defense
2008
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3675524/
————————-
Nanorobotic Applications in Medicine: Current Proposals and Designs
2014
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4562685/
————————
Magnetically guided virus stamping for the targeted infection of single cells or groups of cells
18 October 2019
https://www.nature.com/articles/s41596-019-0221-z
———————————–
Magnets Turn Viruses Into Bacteria-Killers A team of international scientists has used phage-enhanced nanoparticles to kill bacteria that pollute water treatment systems.
Aug. 18, 2017
https://www.asianscientist.com/2017/08/tech/magnet-virus-bacteria-water-treatment/
———————————–
Engineer Finds Way to Pull Diseases From Blood Using Magnets
November 12 2019

“In theory, you can go after almost anything. Poisons, pathogens, viruses, bacteria…”
https://futurism.com/neoscope/engineer-pull-diseases-blood-magnets
———————————-
12 magnets show how viruses are built
Apr 18, 2019
https://www.youtube.com/watch?v=3X6qEE2fHvE
———————————-
Magnets may one day cull deadly germs from blood
February 6, 2017
Tiny particles of antibody-covered iron would grab bacteria, then get mopped up by magnet
https://www.sciencenewsforstudents.org/article/magnets-may-one-day-cull-deadly-germs-blood
———————————-
Biology, genetics, nanotechnology, neuroscience, materials science, biotech, chemical engineering, 3 d, super computing, quantum physics, energy, design, & sustainability
Nov 10, 2014
Genotoxic effect
https://www.slideshare.net/sreeremyasasi/genotoxic-effect
———————————-
Nanorobots that hide in your blood like viruses could someday fight cancer
2014
When it comes to fighting disease, your body’s defense system doesn’t like enlisting outside help. Overcoming this ”locals only” attitude has been a huge handicap for scientists trying to make medical nanorobots, but now a team from Harvard thinks they’ve developed a disguise that will help the nanorobots sneak through and get to work fighting cancer.
The new technology builds structural hangers out of genetic code to attach a fatty covering to the nanodevice. This covering is similar to what some viruses use to move through our bodies undetected.

———————————-
DNA Nanobots Can Fool the Immune System by Disguising Themselves as Viruses
2014
Scientists took a cue from the one of the most efficient genetic-code delivery systems in history or biology.
https://www.vice.com/en_us/article/xywy8a/dna-nanobots-can-fool-the-immune-system-by-disguising-themselves-as-viruses
—————————
‘Nanobot’ viruses tag and round up bacteria in food and water
2018
Tweaking DNA and adding magnetic nanoparticles creates a new tool to test for contaminants
https://www.sciencenews.org/article/nanobot-viruses-tag-and-round-bacteria-food-and-water
—————————-
Future of lung treatment: Malaysian scientists join Harvard team creating safe, effective nano drugs
2016
Additional Malaysian nanoscience research includes converting greenhouse gases into energy source; ‘Smart farming’ nanosensors; New program aims for macro impact in health, energy, environment, agriculture, electronics
https://www.eurekalert.org/pub_releases/2016-01/tca-fol010516.php
————————–
Engineered mRNA-expressed antibodies prevent respiratory syncytial virus infection
2018
The lung is a critical prophylaxis target for clinically important infectious agents, including human respiratory syncytial virus (RSV) and influenza. Here, we develop a modular, synthetic mRNA-based approach to express neutralizing antibodies directly in the lung via aerosol, to prevent RSV infections. First, we express palivizumab, which reduces RSV F copies by 90.8%. Second, we express engineered, membrane-anchored palivizumab, which prevents detectable infection in transfected cells, reducing in vitro titer and in vivo RSV F copies by 99.7% and 89.6%, respectively. Finally, we express an anchored or secreted high-affinity, anti-RSV F, camelid antibody (RSV aVHH and sVHH). We demonstrate that RSV aVHH, but not RSV sVHH, significantly inhibits RSV 7 days post transfection, and we show that RSV aVHH is present in the lung for at least 28 days. Overall, our data suggests that expressing membrane-anchored broadly neutralizing antibodies in the lungs could potentially be a promising pulmonary prophylaxis approach.

aPali is anchored to the membrane and inhibits RSV in cells. a Schematic of aPali anchored to the plasma membrane. b Schematic of aPali and sPali mRNA delivery and expression. c Cells were transfected with vehicle control or 1 μg of either aPali heavy chain only, aPali light chain, or both the heavy and light chain mRNAs. 24 h later, cells were fixed, permeabilized, and stained with a donkey anti-human secondary antibody (white).
https://www.nature.com/articles/s41467-018-06508-3
————————–
Adeno-associated Virus-mediated Transgene Expression in Genetically Defined Neurons of the Spinal Cord
2018
https://www.jove.com/video/57382/adeno-associated-virus-mediated-transgene-expression-genetically
Summary
Intraspinal injection of recombinase dependent recombinant adeno-associated virus (rAAV) can be used to manipulate any genetically labelled cell type in the spinal cord. Here we describe how to transduce neurons in the dorsal horn of the lumbar spinal cord. This technique enables functional interrogation of the manipulated neuron subtype.
—————————-
GSK-3 Inhibitor Promotes Neuronal Cell Regeneration and Functional Recovery in a Rat Model of Spinal Cord Injury
2019
https://www.hindawi.com/journals/bmri/2019/9628065/
—————————-
‘Dancing molecules’ successfully repair severe spinal cord injuries
November 11, 2021
After single injection, paralyzed animals regained ability to walk within four weeks
https://www.sciencedaily.com/releases/2021/11/211111153635.htm
—————————-
New DNA nanorobots successfully target and kill off cancerous tumors
2018
https://techcrunch.com/2018/02/12/new-dna-nanorobots-successfully-targeted-and-killed-off-cancerous-tumors/
—————————
Nanomites from GI Joe Become a Reality
2014
https://www.youtube.com/watch?v=JQpIWU_DHWk
Researchers at Pennsylvania State University have inserted nanomotors inside living human cells, and manipulated them using ultrasound pulses. Advanced versions of these nanomotors could be used in the medical field for targeted therapy and other tasks. The video of the nanomotors inside the cell was taken under 1000x magnification. This is a truly remarkable achievement by PSU researchers.
—————————-
Nanomotors controlled inside living human cells for the first time
2014
https://www.theverge.com/2014/2/16/5417266/nanomotors-controlled-inside-living-human-cells-for-the-first-time

Scientists at Penn State University have successfully controlled tiny nanomotors inside living human cells. Consisting of tiny, rocket-shaped bits of metal, the nanomotors were propelled by ultrasonic waves and steered with magnets. Researcher Tom Mallouk wasn’t afraid to talk up potential future applications, saying that the technology could one day be used “to treat cancer and other diseases by mechanically manipulating cells from the inside.” Once inside a living cell, the nanomotors could pulverize the cell’s contents like an “egg beater” or just break the cell’s membrane, Penn State’s note about the research says, which could allow for targeted attacks on specific cells. More importantly, Mallouk says that the nanomotors were able to move independently of one another, instead of the “whole mass of them going in one direction.”
—————————-
Mechanical Force-Triggered Drug Delivery
2016
https://pubs.acs.org/doi/full/10.1021/acs.chemrev.6b00369?src=recsys
Advanced drug delivery systems (DDS) enhance treatment efficacy of different therapeutics in a dosage, spatial, and/or temporal controlled manner. To date, numerous chemical- or physical-based stimuli-responsive formulations or devices for controlled drug release have been developed. Among them, the emerging mechanical force-based stimulus offers a convenient and robust controlled drug release platform and has attracted increasing attention. The relevant DDS can be activated to promote drug release by different types of mechanical stimuli, including compressive force, tensile force, and shear force as well as indirect formats, remotely triggered by ultrasound and magnetic field. In this review, we provide an overview of recent advances in mechanically activated DDS. The opportunities and challenges regarding clinical translations are also discussed.
——————————
Self-Assembling, Self-Propelling, and Self-Destructive Nanovesicle for Drug Delivery
May 31 2017
https://www.azonano.com/news.aspx?newsID=35603
——————————
Programmable Self-Assembly on Multicomponent Nano-Architectures
Apr 18 2010
https://www.azonano.com/article.aspx?ArticleID=2549
——————————
Progress in Nanomedicine and Medical Nanorobotics
http://www.nanomedicine.com/Papers/ProgressNM06.pdf
CONTENTS1.
Nanotechnology and Nanomedicine……………….. 1
2. Medical Nanomaterials and Nanodevices……………. 3
2.1. Nanopores…………………………… 3
2.2. Artificial Binding Sites and Molecular Imprinting…… 4
2.3. Quantum Dots and Nanocrystals……………… 4
2.4. Fullerenes and Nanotubes…………………. 5
2.5. Nanoshells and Magnetic Nanoprobes………….. 6
2.6. Targeted Nanoparticles and Smart Drugs………… 7
2.7. Dendrimers and Dendrimer-Based Devices……… 10
2.8. Radio-Controlled Biomolecules……………… 11
3. Microscale Biological Robots…………………… 13
3.1. Engineered Viruses…………………….. 13
3.2. Engineered Bacteria…………………….. 14
4. Medical Nanorobotics……………………….. 16
4.1. Early Thinking in Medical Nanorobotics……….. 16
4.2. Nanorobot Parts and Components……………. 16
4.3. Self-Assembly and Directed Parts Assembly……… 23
4.4. Positional Assembly and Molecular Manufacturing…. 29
4.5. Nanorobot Applications Designs and Scaling Studies . . 39
References………………………………. 44
—————————–
Nanorobots With Applications in Medicine
2019
https://www.sciencedirect.com/science/article/pii/B9780128139325000030
—————————–
Nanoprobes in Point of Care Precision Medicine
Multiplexed Detection of MicroRNA Biomarkers Using SERS-Based Inverse Molecular Sentinel (iMS) Nanoprobes
https://vodinh.pratt.duke.edu/research/Nanoprobes%20in%20Point%20of%20Care%20Precision%20Medicine
—————————–
Applications of Nanotechnology in Military Medicine
August 30, 2016
http://internetmedicine.com/2016/08/30/51709/
—————————–
Chapter Thirteen – Architecture and application of nanorobots in medicine
2020
https://www.sciencedirect.com/science/article/pii/B9780128174630000137
—————————–
An army of micro-robots can wipe out dental plaque
Date:April 25, 2019Source:University of PennsylvaniaSummary:A swarm of micro-robots, directed by magnets, can break apart and remove dental biofilm, or plaque, from a tooth. The innovation arose from a cross-disciplinary partnership among dentists, biologists, and engineers.

—————————–
Magnetic nanoparticles could stop blood clot-caused strokes
February 23, 2015
http://www.sciencedaily.com/releases/2015/02/150223122427.htm
—————————–
mRNA “Vaccines,” Eugenics, and the Push for Transhumanism
August 15th, 2021
https://dissidentvoice.org/2021/08/mrna-vaccines-eugenics-and-the-push-for-transhumanism/
—————————–
Will New COVID Vaccine Make You Transhuman?
September 29, 2020
https://thevaccinereaction.org/2020/09/will-new-covid-vaccine-make-you-transhuman/
—————————–
New Vaccines Will Permanently Alter Human DNA
Why is the government so maniacal about injecting vaccines?
May 17, 2016
Consider this article in light of the accelerating push to mandate and enforce vaccination across the planet.
The reference is the New York Times, 3/15/15, “Protection Without a Vaccine.” It describes the frontier of research. Here are key quotes that illustrate the use of synthetic genes to “protect against disease,” while changing the genetic makeup of humans. This is not science fiction:
“By delivering synthetic genes into the muscles of the [experimental] monkeys, the scientists are essentially re-engineering the animals to resist disease.”
“’The sky’s the limit,’ said Michael Farzan, an immunologist at Scripps and lead author of the new study.”
“The first human trial based on this strategy — called immunoprophylaxis by gene transfer, or I.G.T. — is underway, and several new ones are planned.”
“I.G.T. is altogether different from traditional vaccination. It is instead a form of gene therapy. Scientists isolate the genes that produce powerful antibodies against certain diseases and then synthesize artificial versions. The genes are placed into viruses and injected into human tissue, usually muscle.”
Here is the punchline: “The viruses invade human cells with their DNA payloads, and the synthetic gene is incorporated into the recipient’s own DNA. If all goes well, the new genes instruct the cells to begin manufacturing powerful antibodies.”
Read that again: “the synthetic gene is incorporated into the recipient’s own DNA.” Alteration of the human genetic makeup. Permanent alteration.
https://www.infowars.com/new-vaccines-will-permanently-alter-human-dna/
—————————————-
LEAKED – The US Supreme Court ruled that vaccinated people around the world are products of the state, not humans. Due to the mRNA-DNA manipulation in the fatal blow. Once you have the vaccination, you are no longer human. They become trans people owned by the organization.
Sep 22, 2021
https://twitter.com/paulrnelson74/status/1440725229781676034
—————————————-
Supreme Court: Pfizer, Moderna et al. may own your genes once you’re injected with their lab-created mRNA, DNA
April 27, 2021
https://thecovidblog.com/2021/04/27/supreme-court-pfizer-moderna-et-al-may-own-your-genes-once-youre-injected-with-their-lab-created-mrna-dna/
—————————————-
New Supreme Court Decision Rules That cDNA Is Patentable What It Means for Research and Genetic Testing
July 9, 2013
https://blogs.scientificamerican.com/guest-blog/new-supreme-court-decision-rules-that-cdna-is-patentablewhat-it-means-for-research-and-genetic-testing/
In a unanimous decision last month, the Supreme Court ruled that naturally occurring genes are not patentable. But, said the Court, cDNA, a man-made copy of the genetic messenger in cells, is patentable. As a geneticist, I have my own opinions about this ruling. But the potential outcomes are important enough that all members of the public, not just biologists, should be equipped with the knowledge to evaluate it. The ruling may significantly affect patients’ access to genetic testing, and it sets an important precedent for future developments in the biotechnology sector.
The company that applied for these patents is Myriad Genetics. Building on the work of researchers around the world, Myriad identified the location and sequence of two genes that are sometimes mutated in breast cancer, known as BRCA1 and BRCA2 (collectively, BRCA1/2). Myriad filed patents for the genes in 1994 and 1995.
People can have their risk of breast or ovarian cancer assessed by finding out if they have mutations in BRCA1/2. Then, one can use this information to increase preventative care measures, like increased screening, or even having both breasts completely removed (a double mastectomy)—an elective surgery recently made famous by Angelina Jolie.
Myriad Genetics is the primary distributor of the BRCA1/2 test, which costs upwards of $3000. Because Myriad owned the patents on BRCA1/2, it was the only company that could administer the test for cancerous mutations.
The Supreme Court ruled that genes cannot be patented because “natural phenomena” are not patentable. That’s good news for doctors, researchers, and anyone who doesn’t like the idea of a company owning patent rights to pieces of your body. It also opens up BRCA1/2 testing to labs other than Myriad. But, the Court also ruled that cDNA, an edited man-made copy of the gene, can be patented. Ruling that cDNA can be patented will have important consequences for research, including research to discover new disease treatments and create new genetic tests.
Few people outside of biology research have heard of cDNA. In order to understand the critical distinction between DNA and cDNA, some background is necessary. Genes, which are made of DNA, contain the information required to make proteins. DNA is double-stranded, like a ladder. The familiar DNA nucleotides A, C, T, and G each have a complementary partner they always pair with: A always pairs with T, and C with G.
To make protein from DNA, several steps must happen (illustrated in the accompanying schematic). First, the DNA pulls apart into two separate strands and a copy is made. Instead of DNA, this copy is made of RNA. The copy, called pre-RNA, is not identical to template DNA. It’s a complementary copy. Next, that pre-RNA is edited so that only the parts that encode protein (the exons) remain. This exons-only version is called mRNA. The cell then uses the mRNA to assemble proteins.
For scientists, working with RNA is difficult; it is unstable and degrades quickly. So it is sometimes advantageous for researchers to extract mRNA and convert it back to stable DNA. The new DNA that’s created from the mRNA is called cDNA (see DNA to cDNA schematic). Just like pre-RNA is a complementary copy of the DNA template, the cDNA is a complementary copy of the mRNA template. It’s worth mentioning that cDNA can occur naturally; certain viruses can copy mRNA to cDNA (in fact, this is where scientists learned the technique).
cDNA is an edited version of the original gene. The naturally occurring gene contains exons, introns, and other genetic material; the cDNA contains only exons. Those exons are the same exons as the original DNA, and appear in the same order (illustrated in schematic).
Myriad did not create the sequence of the BRCA1/2 cDNA; the sequence is a complementary copy of patients’ mRNA. But, because physically making the cDNA meant the scientists created something new, the Supreme Court held that cDNA was eligible to be patented. This is a bit like taking a copyrighted photograph, cutting several chunks out of the middle, and calling the result a new product that is eligible for copyright. On one hand, the new photograph may have significantly different artistic merits than the original one, and wouldn’t have existed without your intervention. Yet, the existing parts are exactly the same as something that was already there.
Why do Myriad’s patent rights to cDNA matter? There are several reasons. First, cDNA is an important research tool. For example, the edited cDNA sequence, not the longer DNA sequence, is often used to create animal models of diseases. Those models are essential for researching new treatments and cures. Without the licensing to BRCA1/2 cDNA, certain cancer research may be restricted to Myriad. Next, cDNA is critical for developing new diagnostic tests for genetic disorders. Since the BRCA1/2 genes themselves are not patented, it may be possible for other companies to develop new genetic tests— but the patented cDNA will make this process much more difficult. Myriad’s current control over BRCA1/2 testing artificially inflates the cost, because they’ve closed out all of the competitors. Also, without other labs to study the BRCA1/2 mutations and testing methods, there is no way to independently verify the results. This is analogous to getting a diagnosis from a doctor, but then being forbidden to seek a second opinion from anyone else. Finally, the issue is about access to information. By sharing the rich dataset Myriad has collected from patients, collaborative research efforts from many labs could lead to better cancer detection and treatments. It may also help to make testing more affordable.
—————————————-
SUPREME COURT OF THE UNITED STATES
ASSOCIATION FOR MOLECULAR PATHOLOGY ET AL. v. MYRIAD GENETICS, INC., ET AL
April 15, 2013
https://archive.is/aFxbT
—————————————-
SUPREME COURT OF THE UNITED STATES
ASSOCIATION FOR MOLECULAR PATHOLOGY ET AL. v. MYRIAD GENETICS, INC., ET AL
April 15, 2013
https://www.supremecourt.gov/opinions/12pdf/12-398_1b7d.pdf
—————————————-
Vaccinated Trans-Humans
2021
https://streamable.com/7kny9d
—————————————-
Dr. Carrie Madej – Covid Shots, DNA and Transhumanism (Controversial)
May 9, 2021
https://forbiddenknowledgetv.net/dr-carrie-madej-covid-shots-dna-and-transhumanism/
—————————————-
Engineers create nanoparticles that deliver gene-editing tools to specific tissues and organs
October 8, 2020
https://phys.org/news/2020-10-nanoparticles-gene-editing-tools-specific-tissues.html
—————————————-
New tissue engineering process brings laboratory-grown organs one step closer
19 January 2021
Researchers have developed a new technique that that could one day enable us to grow fully functional human organs in the laboratory.
https://www.drugtargetreview.com/news/81080/new-tissue-engineering-process-brings-lab-grown-organs-one-step-closer/
—————————————-
Synthetic tissue can repair hearts, muscles, and vocal cords
November 30, 2021
https://www.sciencedaily.com/releases/2021/11/211130150456.htm
—————————————-
New study outlines additional reasons for optimism related to heart regeneration
September 14, 2021
The Texas Heart Institute has announced that a new study from its prolific Cardiomyocyte Renewal Lab has been published in Circulation Research—a peer-reviewed journal from the American Heart Association that reaches clinical and academic cardiologists, basic cardiovascular scientists, physiologists, cellular and molecular biologists, and cardiovascular pharmacologists. The new study identifies that the Wntless (Wls) gene plays a critical role in heart regeneration in mice. Wls facilitates signal molecule secretion from cardiomyocytes (heart muscle cells) to cardiac fibroblasts—cells that produce connective tissue—in order to suppress CF activation, which in turn promotes heart functional recovery by reducing scar formation.

Wls knock-down by CRISPR/Cas9 in cardiomyocytes promotes cardiac fibroblast proliferation during neonatal heart regeneration. Green indicates Vimentin-positive cardiac fibroblasts; red indicates proliferating cells with EdU labeling; blue indicates nuclei by DAPI staining.
—————————————-
Nanomedicine: Addressing Cardiovascular Disease and Cardiovascular Tissue Regeneration
2009
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5019576/
—————————————-
Mapping Cells in the “Immortal,” Regenerating Hydra
July 25, 2019

Hydra boast stem cells that exist in a continuous state of renewal and seem to hold within their genomic code the key to biological immortality.
Unlike the stem cells of an adult human, the stem cells of an adult Hydra—a small freshwater invertebrate related to jellyfish and corals—are in a constant state of renewal, bestowing it with amazing regenerative capabilities and nearly biological immortality. Around 100,000 cells make up the Hydra body, and amazingly, these cells renew every 20 days thanks to the Hydra’s bottomless well of stem cells.
But how do these stem cells produce specialized cells in the animal, like neurons or skin cells? What genetic decision trees do Hydra stem cells use?
In a study appearing in Science, Assistant Professor Celina Juliano, Department of Molecular and Cellular Biology, and her colleagues used single-cell sequencing techniques to explore the genetic trajectory for nearly 25,000 Hydra cells. The study was led by Stefan Siebert, a project scientist in the Juliano Lab, in collaboration with Jeff Farrell, a postdoc at Harvard University.
“The beauty of single-cell sequencing and why this is such a big deal for developmental biologists is that we can actually capture the genes that are expressed as cells differentiate from stem cells into their different cell types,” said Juliano.
The study gives developmental biologists a high resolution “single-cell molecular map” of the three stem cell developmental lineages in Hydra.
Hydra “holds an informative position in the animal tree of life,” said Siebert. “Data sets like ours will help us to better understand important gene regulatory networks that were in place early in evolution and that are shared among animals and us.”
The research also provides the first gene expression map of the Hydra nervous system. Understanding how the Hydra regenerates its entire nervous system could help us better understand neurodegenerative diseases in humans and open the door to new therapies.
The three fountains of eternal youth
Hydra continuously renew their cells from three different stem cell populations, which are known as endodermal epithelial, ectodermal epithelial and interstitial cells. To trace the differentiation of individual stem cells, Juliano and her colleagues used a technique called Drop-seq, which was developed by the McCarroll Lab at Harvard University.
Using Hydra cells, barcoded beads and microfluidic devices, the researchers analyzed sets of messenger RNA molecules, called transcriptomes, from individual Hydra cells and grouped them together by cell type based on the genes each cell expresses.
“Single-cell sequencing is a game-changing technology and gives us access to information that is usually masked when sequencing whole tissues,” said Siebert. “Drop-seq is a very cost-efficient version of this type of protocol allowing us to analyze the cells of a whole organism.”
Juliano and her colleagues started Drop-seq runs on Hydra cells more than two years ago. An empty, celebratory champagne bottle sits on a shelf in Juliano’s office and marks the date of the lab’s first Drop-seq run, Feb. 24, 2017. Farrell wrote on the bottle, “Not a drop left.”

Unlike the stem cells of an adult human, the stem cells of an adult Hydra are in a constant state of renewal
https://biology.ucdavis.edu/news/mapping-cells-immortal-regenerating-hydra
—————————————-
Cellular and Molecular Mechanisms of Hydra Regeneration
2019
https://pubmed.ncbi.nlm.nih.gov/31598861/
—————————————-
Components, structure, biogenesis and function of the Hydra extracellular matrix in regeneration, pattern formation and cell differentiation
2012
https://pubmed.ncbi.nlm.nih.gov/22689358/
—————————————-
Real time dynamics of β-catenin expression during Hydra development, regeneration and Wnt signalling activation
2018
https://pubmed.ncbi.nlm.nih.gov/29877570/
—————————————-
Hydra as a unique model for the study of regenerative mechanisms in metazoans
September 2019
https://medcraveonline.com/MOJAP/MOJAP-06-00264.pdf
—————————————-
The Hydra model – a model for what?
February 2012

Regeneration of Hydra from aggregated cells. Hydra cells form random aggregates; ejected from a pipette (A), they sort out to generate a hollow structure with ectoderm and endoderm typical of Hydra tissue (B). Then, new heads are formed, grow out (C) and eventually separate into normal polyps. Such regeneration is prototypical for de novo pattern formation in development and its modelling by theoretical analysis.
https://www.researchgate.net/publication/221982519_The_Hydra_model_-_a_model_for_what
—————————————-
The Hydra model: disclosing an apoptosis-driven generator of Wnt-based regeneration
June 21, 2010
https://www.cell.com/trends/cell-biology/fulltext/S0962-8924(10)00099-1#relatedArticles
—————————————-
What is Regeneration?
2019
What is Regeneration?
Regeneration is one of the processes in which if an organism is cut into several pieces, each of its parts regrows to the original state. This process is carried out by specialized cells called stem cells. It takes place in organisms that have a very simple structure with very few specialized cells.
The cells divide quickly into a large number of cells. Each cell undergoes changes to form various cell types and tissues. This sequential process of changes is known as development. The tissues form various body parts and organs.
Types of Regeneration
There are two types of regeneration:
Morphallaxis
This type of regeneration has little growth and depends upon tissues repatterning. For eg., Hydra grows by loss of cells from its end and by budding. When cut into two, the upper part develops into foot while the lower part develops into the head. The formation of nearby heads is inhibited by the head region.
Epimorphosis
This type of regeneration depends upon the growth of new and properly patterned structures. For eg., regeneration of a vertebrate limb. It involves cell dedifferentiation and growth. The epidermal cells form a blastema. The blastema gives rise to structures with positional distal values. Retinoic acid changes proximal-distal values in limb regeneration.
Which Organisms can Regenerate?
Regeneration occurs in organisms like hydra, flatworms, tapeworms. They have highly adaptive regenerative capabilities. When an organism is wounded, its cells are activated and the damaged tissues and organs are remodelled back to the original state.
Regeneration is very prominent among metazoans. Starfish, crayfish, reptiles, and amphibians also exhibit signs of tissue regeneration. It is not the same as reproduction. In some animals such as the lizard, the shed limb regrows into the original organ.
Regeneration can happen in many different ways using pluripotent stem cells. Some regeneration does not require stem cells. After amputation, stem cells accumulate at the site of injury. The cells then start dividing to form the missing tissue. But not all animals use the pluripotent cells for regeneration.
Regeneration in Plants
Plants can regenerate all body part from the precursor cells. For eg., several trees are cut at the bottom. After some time sprouts appear at the margin of the stump that develop into new leaves, flowers, and stems.
The entire plant can be developed by callus differentiation in the laboratory through plant tissue culture.
Regeneration in Hydra
Hydra had been used as the model of regeneration since time immemorial. Once wounded their cells start to regenerate and grow into pre-existing cells.
Hydra grows by continuously losing cells from its base and by budding
↓
A gradient of inhibitor is produced by the head. This prevents the formation of other heads
↓
The concentration of inhibitor is reduced
↓
The positional value increases locally
↓
A new head is formed without any new growth

—————————————-
“Immortal” Hydra Vulgaris In Moderna’s Vaccine? Dr Carrie Madej’s Startling Microscope Findings & Up
October 4th, 2021
https://www.bitchute.com/video/YBwNSE7b6nyy/
—————————————-
Team rewires a behavioral circuit in a worm using hydra parts
September 29, 2021

C. elegans worms with green fluorescent protein (GFP) inserted into their neurons to visualize neural development.
For two people to communicate in a loud, crowded room, they need to be standing side by side. The same is often true for neurons in the brain. But the same way a cell phone allows two people to communicate clearly across the room, new research at the Marine Biological Laboratory (MBL) has opened up a new channel of communication in the brain of the worm C. elegans.
The research, published in Nature Communications, highlights the development of HySyn, a system designed to synthetically reconnect neural circuits using neuropeptides from Hydra, a small, freshwater organism. (Neuropeptides modulate the activity of neurotransmitters to increase or decrease the strength of impulses between neurons.)
For the first time, the researchers created genetic lines of mutant C. elegans that expressed neuropeptides from the Hydra brain, creating an artificial synapse that rewired a behavioral circuit in the worm. Because none of the other synapses in the brain, besides those fitted with the hydra receptor and neuropeptide, could hear the “command,” it was like giving them a cell phone so they could communicate.
“These neuromodulatory peptides let you communicate at a distance,” said MBL Fellow Daniel Colón-Ramos of Yale University School of Medicine. “It gives you more flexibility as a researcher to manipulate neurons that are not adjacent to each other.” Colón-Ramos, senior author on the paper, was postdoctoral advisor for the paper’s first author, former MBL Grass Fellow Josh Hawk. The work and analysis was performed at the MBL and at Yale University in Colón-Ramos’s lab.
The researchers used a mutant line of C. elegans that was missing the neural connection that controlled specific behavior—the behavior that told them that they were full and needed to stop searching for food. By taking genes that encode a neuropeptide and its receptor from Hydra and putting them into the C. elegans worm, researchers were able to restore the neural circuit that controls this behavior. They created two separate genetic lines—one that contained the neuropeptide and one that contained the receptor. The offspring of the pair contained the full neural peptide pathway. But, according to Hawk, it’s just one possible pathway to focus on. “There are hundreds of neural peptides in Hydra, each of which could be a different channel of communication,” said Hawk. “To me, that’s the most exciting thing. This should open up a whole area that no one has ever explored before.
—————————————-
Low-voltage magnetoelectric coupling in membrane heterostructures
November 19, 2021
https://phys.org/news/2021-11-low-voltage-magnetoelectric-coupling-membrane-heterostructures.html
—————————————-
Repairing DNA damage in the human body
April 13, 2016
https://www.sciencedaily.com/releases/2016/04/160413135701.htm
—————————————-
How to modify RNA: Crucial steps for adding chemical tag to transfer RNA revealed
September 15, 2021
https://phys.org/news/2021-09-rna-crucial-adding-chemical-tag.html
—————————————-
A brief history of the study of nerve dependent regeneration
17 Apr 2017
https://www.tandfonline.com/doi/pdf/10.1080/23262133.2017.1302216
—————————————-
Alternative Regenerative Medicine for Diabetes: Beyond the Stem Cell Approach
https://www.frontiersin.org/research-topics/8598/alternative-regenerative-medicine-for-diabetes-beyond-the-stem-cell-approach
—————————————-
The role of stem cells in limb regeneration.
01 Jan 2016
http://europepmc.org/article/PMC/4882123
—————————————-
Stem Cells Help Lizard Regenerate a Perfect Tail for First Time in More Than 250 Million Years
October 25, 2021
https://scitechdaily.com/stem-cells-help-lizard-regenerate-a-perfect-tail-for-first-time-in-more-than-250-million-years/?utm_source=TrendMD&utm_medium=cpc&utm_campaign=SciTechDaily_TrendMD_0
—————————————-
Humans have the salamander-like ability to regenerate…
2019

Much work is being done on human limb regeneration concentrating on the specific microRNA circuit used by zebrafish, bichirs, and axolotls
https://www.pinterest.com/pin/444519425723791938/
—————————————-
Salamander genome gives clues about unique regenerative ability
Dec 22, 2017
https://news.cision.com/karolinska-institutet/r/salamander-genome-gives-clues-about-unique-regenerative-ability,c2420608
—————————————-
Regrowing limbs: fossils reveal ancient secrets of salamander ancestors
October 26, 2015
https://theconversation.com/regrowing-limbs-fossils-reveal-ancient-secrets-of-salamander-ancestors-48999
—————————————-
Salamander and Regeneration Science
Jul 9, 2019
Why does PainScience.com have a salamander mascot? Their regenerative superpower is an inspiring, profound example of what is possible in biology and healing
Other kinds of critter regeneration
Many tiny organisms are masters of regeneration. But it’s rare in animals larger than a speck, and particularly rare in mammals. If regeneration is possible in any mammal, then there’s hope for us.
And it is possible. This neat 2012 science story is promising: “Biologist discovers mammal with salamander-like regenerative abilities.”4 The African spiny mouse does a far better job at regenerating any part of itself than any other known mammal to date. Salamanders are much better at regeneration, in every way, but at least we know mammals aren’t completely left out of the regeneration game.
And there are many other examples of limited critter regeneration of specific body tissues and parts.
Another mouse may deserve even more attention than the African spiny: the MRL mouse is a strain of mouse that exhibits diverse regenerative abilities. While they fall short of the salamander, they heal from all kinds of wounds far better than we do.
The ogre-faced spider has better vision that we do, and it never gets old — its eyes specifically, literally, never get old, because the O-F spider replaces its retinas every night.5
Many species can shed limbs and tails parts as a defense and then regrow them (automy).
The annelids (flatworms) are unusually good at regeneration. For instance, a planarian split lengthwise or crosswise will regenerate into two separate individuals. Quite a party trick.
The mole rat’s superpower
“To deal with their miserable lives” naked mole rats evolved to feel no pain:
And yet in this harsh environment, under extremely crowded conditions, the naked mole rat has evolved to be virtually indestructible: these small mammals almost never get cancer, live to be over 30 (much longer than other rat species), and they are insensitive to acid burns. A study in Cell Reports reveals one secret behind these rats’ abilities. Evolutionary tweaks to the amino acids in their pain receptors make naked mole rats extremely insensitive to pain after they are born.
I think it’s surprising and fascinating that immunity to pain isn’t a more common adaptation in biology. Clearly pain has potent survival benefits — the ultimate double-edged sword.
https://www.painscience.com/about-salamander.php
—————————————-
Novel insights into the flexibility of cell and positional identity during urodele limb regeneration
2008 Nov 21
https://pubmed.ncbi.nlm.nih.gov/19028989/
—————————————-
Amphibian Limb Regeneration: Rebuilding a Complex Structure
4 Apr 1997
https://www.science.org/doi/10.1126/science.276.5309.81
—————————————-
Glial Cell Behavior Critical to Proficient Central Nervous System Regeneration
April 24th, 2018
https://www.fightaging.org/archives/2018/04/glial-cell-behavior-critical-to-proficient-central-nervous-system-regeneration/
Why can species such as salamanders regrow organs and limbs while mammals cannot? This proficiency even extends to portions of the central nervous system, such as the spinal cord. In recent years, researchers have made good progress in understanding exceptional regeneration, finding that, for example, differences in the behavior of immune cells called macrophages are essential to regrowth. In the central nervous system, glial cells are somewhat analogous to macrophages in other tissues, and in the research noted here, scientists report on evidence for an equivalent importance in mammalian versus salamander regenerative capacities.
Given the macrophage and glial cell connection, this area of comparative biology is moving of late from speculative to relevant to clinical development. Numerous research groups are investigating the alteration of macrophage and glial cell behavior in order to spur greater regeneration in mammals. These cells can be classified by their behavior, either aggressive and inflammatory while seeking out pathogens, or more focused on aiding regeneration. Both behaviors are needed, but in mammals, and in the old, there is too much of the first type and too little of the second type of behavior. In learning to adjust cell behavior to change this imbalance, the foundations may be laid for more profound enhancements of regeneration in the years ahead, building on what is learned from salamanders.
—————————————-
Cell proliferation controls body size growth, tentacle morphogenesis, and regeneration in hydrozoan jellyfish Cladonema pacificum
August 26, 2019
https://peerj.com/articles/7579/
—————————————-
Whole-Body Regeneration in Sponges: Diversity, Fine Mechanisms, and Future Prospects
2021 Mar 29
https://www.ncbi.nlm.nih.gov/labs/pmc/articles/PMC8066720/
—————————————-
The cellular and molecular bases of the sponge stem cell systems underlying reproduction, homeostasis and regeneration.
21 June 2018
https://www.semanticscholar.org/paper/The-cellular-and-molecular-bases-of-the-sponge-stem-Funayama/4e5e6ad2191abd87db0e99ae23a403cc54d08366
—————————————-
Cell kinetics during regeneration in the sponge Halisarca caerulea: how local is the response to tissue damage?
2015
https://peerj.com/articles/820.pdf
—————————————-
Regeneration in bipinnaria larvae of the bat star Patiria miniata induces rapid and broad new gene expression
2016
https://www.brown.edu/research/labs/wessel/sites/wessel/files/Oulhen%20et%20al.%2C%202016%20regeneration.pdf
—————————————-
From One to Many and Back Again: A Systemic Signal Triggers Tunicate Regeneration
March 6, 2007
For a lucky subset of vertebrates, losing an appendage is no big deal. As many an inquisitive child knows, salamanders can regenerate lost limbs or tails; and as lab investigators know, zebrafish can regrow lost fins. Of course, humans and other “higher” vertebrates must make do with repairing rather than regenerating damaged tissues. Though whole body generation (WBR) does occur, it’s typically restricted to a subset of morphologically less complex invertebrates, such as sponges, flatworms, and jellyfish.
In a new study, Yuval Rinkevich et al. shed light on the molecular signals underlying WBR by investigating the phenomenon in our closest invertebrate relative, the sea squirt Botrylloides leachi. The researchers identified several novel features of WBR in this colonial marine organism, and discovered an unusual mode of regeneration.
As invertebrate members of the phylum Chordata, sea squirts share several fundamental biological pathways with vertebrates; consequently, using them as a model system to study WBR could illuminate not only the evolutionary origin of regeneration, but also its subsequent attenuation in vertebrates.
Sea squirts (also called “tunicates” after their tough outer tunic) are widely distributed in shallow coastal waters as colonies of genetically identical individuals called “zooids.” These clones develop from sexually reproduced larvae that adhere to rocks or other substrates on the sea floor soon after hatching. Once attached, the tadpole-like larvae lose several of the features that make them chordates—the notochord, dorsal nerve tube, and postanal tail (muscle tissue beyond the digestive tract)—and develop adult organs, including a digestive tract equipped with pharyngeal slits, endostyle, neural complex, heart, and gonads.
Thus transformed, the sedentary tunicate initiates the rounds of asexual reproduction that create the colony. During this four-stage process, called blastogenesis (or palleal budding), new buds sprout from the thoracic body wall of the founding “oozooid,” spawning zooids that repeat the process. As new buds mature, parental zooids degrade and undergo resorption into the colony. Zooids connect to colony mates through a network of blood vessels with delicate fingerlike projections called ampullae. Experiments in a close relative of B. leachi showed that buds forming at the base of these vascular ampullae supported WBR.
To investigate WBR in B. leachi, Rinkevich et al. collected colonies from the Mediterranean coast of Israel and analyzed the morphological, cellular, and molecular characteristics of the process. The researchers removed fragments of blood vessels with ampullae from the colonies, and placed the fragments on slides for regeneration. Of 95 fragments, 80 underwent WBR.
Following dissection, vessels within fragments contracted and blood flow abated; within a day, a new circulatory system emerged as vascular connections between ampullae grew and blood flow returned. Ampullae dynamically changed shape and position over the course of a week, creating a dense localized network with new blood vessels opposite a vessel-free gelatinous tunic matrix. Blood flow attenuated, and an opaque mass of vessels formed around a transparent vesicle with two openings. Within 10 to 14 days, this opaque mass yielded functional siphons (an inhalant, or peribranchial, siphon and an exhalant, or atrial, siphon) and a “fully operating filter-feeding zooid.” Among several regeneration starts, only the fastest developing bud in a fragment reached the final zooid stage; the others were absorbed into the colony.
Each day, Rinkevich et al. placed regenerating fragments under the microscope to study their cellular changes. By the second day, dozens of small compartments—the newly dubbed regeneration niches—started forming and filling with clusters of blood cells. Over the next few days, aggregating cells formed around a hollow sphere, then reorganized into a thin and thick layer on opposite sides, very similar to early stages of embryonic development. As cells proliferated, buds grew, and the thick cell layer folded inward, forming double-walled folds (which became the pharyngeal slits) and creating a middle chamber and two side chambers (which became the pharyngeal chamber and lateral atrial chambers). Organ development continued, and an adult zooid, capable of sexual reproduction, appeared within two weeks.
For molecular insights into regeneration, the researchers focused on retinoic acid (RA) signaling by examining the temporal expression of its receptor (RAR). In addition to its role in chordate body patterning, RA (a vitamin A metabolite) induces the regeneration of several tissues and organs. Only regenerating vessels and ampullae expressed RAR, and this expression continued through each phase of regeneration.
The researchers confirmed RA’s vital role in regeneration by inhibiting RA synthesis with chemicals and destroying RA transcripts with RNA interference. In both cases, malformed buds failed to generate zooids from dissected fragments. Similar problems occurred when RAR function was disrupted. In contrast, RA overexpression led to accelerated regeneration, with multiple buds reaching the fully developed zooid stage. RAR regulates developmental elements of the normal budding process in a sister colonial tunicate species, suggesting that organisms recruit the same signals for development and regeneration.
In regenerating fully functional adult tunicates from “minute vascular fragments,” the researchers identified several features of this system that differ from those of established regeneration model systems. In contrast to limb or fin regeneration—which arises from local signals emanating from a “regeneration center”—B. leachi WBR arises from systemically induced signals in multiple “regeneration niches.” These niches arise from the vascular network (rather than from proliferating balls of cells), and regeneration appears to be regulated by systemic (rather than local) cues. These systemic cues, the researchers propose, may travel through the circulation, thereby supporting multiple regeneration foci. The researchers plan to investigate the cellular source of the tunicate’s remarkable regenerative power in future studies.
https://journals.plos.org/plosbiology/article?id=10.1371/journal.pbio.0050098
—————————————-
Integrin-alpha-6+ Candidate stem cells are responsible for whole body regeneration in the invertebrate chordate Botrylloides diegensis
07 September 2020
https://www.nature.com/articles/s41467-020-18288-w
—————————————-
Whole body regeneration and developmental competition in two botryllid ascidians.
15 Dec 2021
https://europepmc.org/article/PMC/PMC8675491
—————————————-
Systemic Bud Induction and Retinoic Acid Signaling Underlie Whole Body Regeneration in the Urochordate Botrylloides leachi
https://citeseerx.ist.psu.edu/viewdoc/summary?doi=10.1.1.278.9480
—————————————-
Pattern of Cell Proliferation During Budding in the Colonial Ascidian Diplosoma listerianum
2010
https://www.journals.uchicago.edu/doi/full/10.1086/BBLv221n1p126
—————————————-
Telomerase maintained in self-renewing tissues during serial regeneration of the urochordate Botryllus schlosseri
2004
https://pubmed.ncbi.nlm.nih.gov/15328006/
—————————————-
Elucidating the role of apoptosis during cyclical body regeneration in Botryllus schlosseri
6-2013
https://digitalworks.union.edu/cgi/viewcontent.cgi?article=1652&context=theses
—————————————-
Telomerase deficiency in a colonial ascidian after prolonged asexual propagation
2011
https://pubmed.ncbi.nlm.nih.gov/21548077/
—————————————-
Asexual Propagation and Regeneration in Colonial Ascidians
2011
https://www.journals.uchicago.edu/doi/full/10.1086/BBLv221n1p43
—————————————-
Zooid
Comparative Reproduction
Fabio Gasparini, Loriano Ballarin, in Encyclopedia of Reproduction (Second Edition), 2018
Pyrosomatida
Pyrosomes, common in tropical and temperate seas, owe their name to luminescent organs which contain symbiotic bioluminescent bacteria. These bacteria enter the eggs and are transmitted to the new colonies. Colonies have the form of hollow cylinders with one blind end
Their size ranges from few centimetres to more than one metre in length. Zooids are located in the wall of the cylinder, embedded in the common tunic; the oral siphon of each zooid faces outwards, whereas the atrial aperture opens into the central cloacal cavity: water expelled through the cloacal aperture allows colonial locomotion by propulsion. The general structure of pyrosome zooids is quite similar to that of their ascidian counterparts: they are, hermaphroditic (an ovary and a testis lying ventrally, near the gut) and have a wide branchial basket occupying most of the body volume (Fig. 6).
Colonies grow by addition of new zooids that bud from a short stolon in the heart region of parental zooids. In a colony the oldest zooids are protandric and located near to the closed end; conversely, younger zooids are protogynous and located near the colonial opening, those in the middle are simultaneously hermaphroditic. Self-fertilisation is possible. Eggs, surrounded by both test and follicular cells, are provided with abundant yolk. Fertilisation is internal, and the zygote undergoes partial cleavage in a peribranchial pouch. A tadpole larva is missing and, at the completion of development, a goblet-like oozooid, named cyatozooid, is released from the atrial siphon. A stolon originates from its endostylar pharyngeal wall that buds four primary blastozooids. At this point the new colony reaches superficial waters and grows by repeated cycles of asexual reproduction.
https://www.sciencedirect.com/topics/immunology-and-microbiology/zooid
—————————————-
Chapter Eight – Tunicate gastrulation
2020
Abstract
Tunicates are a diverse group of invertebrate marine chordates that includes the larvaceans, thaliaceans, and ascidians. Because of their unique evolutionary position as the sister group of the vertebrates, tunicates are invaluable as a comparative model and hold the promise of revealing both conserved and derived features of chordate gastrulation. Descriptive studies in a broad range of tunicates have revealed several important unifying traits that make them unique among the chordates, including invariant cell lineages through gastrula stages and an overall morphological simplicity. Gastrulation has only been studied in detail in ascidians such as Ciona and Phallusia, where it involves a simple cup-shaped gastrula driven primarily by endoderm invagination. This appears to differ significantly from vertebrate models, such as Xenopus, in which mesoderm convergent extension and epidermal epiboly are major contributors to involution. These differences may reflect the cellular simplicity of the ascidian embryo.
https://www.sciencedirect.com/science/article/abs/pii/S0070215319300742
—————————————-
Gastrulation: From Embryonic Pattern to Form
2020
4.1 Colonial tunicates
A subset of ascidian species are colonial and grow by repeated rounds of asexual reproduction (budding) that eventually give rise to a colony of genetically identical individuals, called zooids, arranged within a common tunic. Each zooid can be sexually mature, but colonial reproduction has several notable differences when compared to solitary species, and these have hampered embryological studies. First, all colonial ascidians are brooders, and fertilization and development occur inside the adult zooids, often with maternal contributions, such as a brood pouch, which can be difficult to replicate in vitro. Development is also significantly slower, on the order of days to a week or more. Finally, the eggs and developing embryos are opaque, hindering high-resolution studies of cell movement. Thus little is known about gastrulation in these species, and there are no recently published studies.
More germane to this review is the potential role of a gastrulation-like process during asexual development. During the budding process, colonial ascidians are regenerating all tissues and organs from a population of pluripotent stem cells. An important question is whether asexual development is controlled by novel genes and gene regulatory networks, or if embryonic pathways and networks are redeployed (Tiozzo, Brown, & De Tomaso, 2008). Interestingly, there are many colonial species scattered throughout the class Ascidiacea, including different monophyletic orders, suggesting that coloniality has arisen independently multiple times. Comparison of these species reveals a diversity of budding modes, including the source of the new body, and the timing of various developmental landmarks (Tiozzo et al., 2008). However, in all species studied to date, asexually developing zooids go through an initial blastula-like stage. This is followed by invaginations and evaginations of the epithelium to form various tissues and organs. While in a sense this resembles gastrulation, these movements do not result in segregation of cells into presumptive germ layers, as they do in embryogenesis. The only species in which asexual budding has been studied at a molecular level is Botryllus schlosseri (Manni et al., 2019). In an elegant study by Ricci, Cabrera, Lotito, and Tiozzo (2016), it was found that there is regionalization of germ layer specific transcription factors in the blastula-like stage and during these epithelial folding events, but they do not correspond to the regions that are folding (i.e., it does not appear that the movement is correlated to germ layer specification). In this study, it was found that germ layer restricted transcription factors, such as Otx (ectoderm), Fox-A1 (endoderm) and Gsc (mesoderm) are expressed in distinct regions of the blastula-like structure and could be followed through organogenesis. Importantly, the expression of each of these germ layer markers corresponded to the source of mature organs during embryogenesis. For example, the endoderm marker Fox-A1 was initially expressed in a region of the blastula-like vesicle that eventually became the gut, and results were equivalent for ectodermal and mesodermal markers.
Given that coloniality has arisen multiple times, it would hardly be feasible that completely novel mechanisms for regeneration of every tissue and organ also evolved multiple times, and a more parsimonious explanation is that this is due to the ability to coopt embryonic pathways at different times during regeneration. Interestingly though, this does not appear to include a clear recapitulation of embryonic gastrulation.
https://www.sciencedirect.com/topics/immunology-and-microbiology/zooid
—————————————-
Mitochondrial ribosome
The mitochondrial ribosome, or mitoribosome, is a protein complex that is active in mitochondria and functions as a riboprotein for translating mitochondrial mRNAs encoded in mtDNA. The mitoribosome is attached to the inner mitochondrial membrane. Mitoribosomes, like cytoplasmic ribosomes, consist of two subunits — large (mtLSU) and small (mt-SSU). Mitoribosomes consist of several specific proteins and fewer rRNAs. While mitochondrial rRNAs are encoded in the mitochondrial genome, the proteins that make up mitoribosomes are encoded in the nucleus and assembled by cytoplasmic ribosomes before being implanted into the mitochondria.
https://en.wikipedia.org/wiki/Mitochondrial_ribosome
—————————————-
Ribosome
Ribosomes ( /ˈraɪbəˌsoʊm, -boʊ-/), also called Palade granules (after discoverer George Palade and due to their granular structure), are macromolecular machines, found within all cells, that perform biological protein synthesis (mRNA translation). Ribosomes link amino acids together in the order specified by the codons of messenger RNA (mRNA) molecules to form polypeptide chains. Ribosomes consist of two major components: the small and large ribosomal subunits. Each subunit consists of one or more ribosomal RNA (rRNA) molecules and many ribosomal proteins (RPs or r-proteins). The ribosomes and associated molecules are also known as the translational apparatus.
https://en.wikipedia.org/wiki/Ribosome
—————————————-
Ribozyme
https://en.wikipedia.org/wiki/Ribozyme
Ribozymes (ribonucleic acid enzymes) are RNA molecules that have the ability to catalyze specific biochemical reactions, including RNA splicing in gene expression, similar to the action of protein enzymes. The 1982 discovery of ribozymes demonstrated that RNA can be both genetic material (like DNA) and a biological catalyst (like protein enzymes), and contributed to the RNA world hypothesis, which suggests that RNA may have been important in the evolution of prebiotic self-replicating systems. The most common activities of natural or in vitro-evolved ribozymes are the cleavage or ligation of RNA and DNA and peptide bond formation. For example, the smallest ribozyme known (GUGGC-3′) can aminoacylate a GCCU-3′ sequence in the presence of PheAMP. Within the ribosome, ribozymes function as part of the large subunit ribosomal RNA to link amino acids during protein synthesis. They also participate in a variety of RNA processing reactions, including RNA splicing, viral replication, and transfer RNA biosynthesis. Examples of ribozymes include the hammerhead ribozyme, the VS ribozyme, Leadzyme and the hairpin ribozyme.
Researchers who are investigating the origins of life through the RNA world hypothesis have been working on discovering a ribozyme which has the capacity to self-replicate, which would require it to have the ability to catalytically synthesize polymers of RNA. This should be able to happen in prebiotically plausible conditions with high rates of copying accuracy to prevent degradation of information but also allowing for the occurrence of occasional errors during the copying process to allow for Darwinian evolution to proceed.
Attempts have been made to develop ribozymes as therapeutic agents, as enzymes which target defined RNA sequences for cleavage, as biosensors, and for applications in functional genomics and gene discovery.
—————————————-
HDV-like self-cleaving ribozymes
2011 Sep 1
https://www.ncbi.nlm.nih.gov/labs/pmc/articles/PMC3256349/
HDV ribozymes catalyze their own scission from the transcript during rolling circle replication of the hepatitis delta virus. In vitro selection of self-cleaving ribozymes from a human genomic library revealed an HDV-like ribozyme in the second intron of the human CPEB3 gene and recent results suggest that this RNA affects episodic memory performance. Bioinformatic searches based on the secondary structure of the HDV/CPEB3 fold yielded numerous functional ribozymes in a wide variety of organisms. Genomic mapping of these RNAs suggested several biological roles, one of which is the 5′ processing of non-LTR retrotransposons. The family of HDV-like ribozymes thus continues to grow in numbers and biological importance.
—————————————-
The identification and characterization of a selected SAM-dependent methyltransferase ribozyme that is present in natural sequences
20 October 2021
https://www.nature.com/articles/s41929-021-00685-z
—————————————-
Small circRNAs with self-cleaving ribozymes are highly expressed in diverse metazoan transcriptomes
21 March 2020
https://academic.oup.com/nar/article/48/9/5054/5810855?login=false
—————————————-
Beyond Adult Stem Cells: Dedifferentiation as a Unifying Mechanism Underlying Regeneration in Invertebrate Deuterostomes
2020 Oct 20
https://www.ncbi.nlm.nih.gov/labs/pmc/articles/PMC7606891/
—————————————-
The Nervous System is regenerated in Sea Star Larvae Through Reuse of the Embryonic Neural Stem Cell Lineage and Re-activation of Stem Cell Specification
29.08.2019
https://kilthub.cmu.edu/articles/thesis/The_Nervous_System_is_regenerated_in_Sea_Star_Larvae_Through_Reuse_of_the_Embryonic_Neural_Stem_Cell_Lineage_and_Re-activation_of_Stem_Cell_Specification/9584741
—————————————-
Molecular mechanisms of wound healing and regeneration of siphon in the Manila clam Ruditapes philippinarum revealed by transcriptomic analysis
2021
https://www.sciencedirect.com/science/article/abs/pii/S0888754321000707
—————————————-
The Cnidarian and its Regenerative Properties
https://www.coursehero.com/file/14045426/Cnidarian-Regeneration/
—————————————-
Reproductive mode, stem cells and regeneration in a freshwater cnidarian with postreproductive senescence
24 July 2018
https://besjournals.onlinelibrary.wiley.com/doi/10.1111/1365-2435.13189
—————————————-
PdumBase: a transcriptome database and research tool for Platynereis dumerilii and early development of other metazoans
16 August 2018
https://bmcgenomics.biomedcentral.com/articles/10.1186/s12864-018-4987-0
—————————————-
Drosophila as a Model System to Study Cell Signaling in Organ Regeneration
19 Mar 2018
Abstract
Regeneration is a fascinating phenomenon that allows organisms to replace or repair damaged organs or tissues. This ability occurs to varying extents among metazoans. The rebuilding of the damaged structure depends on regenerative proliferation that must be accompanied by proper cell fate respecification and patterning. These cellular processes are regulated by the action of different signaling pathways that are activated in response to the damage. The imaginal discs of Drosophila melanogaster have the ability to regenerate and have been extensively used as a model system to study regeneration. Drosophila provides an opportunity to use powerful genetic tools to address fundamental problems about the genetic mechanisms involved in organ regeneration. Different studies in Drosophila have helped to elucidate the genes and signaling pathways that initiate regeneration, promote regenerative growth, and induce cell fate respecification. Here we review the signaling networks involved in regulating the variety of cellular responses that are required for discs regeneration.
https://www.hindawi.com/journals/bmri/2018/7359267/
—————————————-
Klf5 establishes bi-potential cell fate by dual regulation of ICM and TE specification genes
2021
https://www.biorxiv.org/content/10.1101/2021.06.02.446799v1
—————————————-
Regeneration in the ctenophore Mnemiopsis leidyi occurs in the absence of a blastema, requires cell division, and is temporally separable from wound healing
11 October 2019
The ability to regenerate is a widely distributed but highly variable trait among metazoans. A variety of modes of regeneration has been described for different organisms; however, many questions regarding the origin and evolution of these strategies remain unanswered. Most species of ctenophore (or “comb jellies”), a clade of marine animals that branch off at the base of the animal tree of life, possess an outstanding capacity to regenerate. However, the cellular and molecular mechanisms underlying this ability are unknown. We have used the ctenophore Mnemiopsis leidyi as a system to study wound healing and adult regeneration and provide some first-time insights of the cellular mechanisms involved in the regeneration of one of the most ancient extant group of multicellular animals.
https://bmcbiol.biomedcentral.com/articles/10.1186/s12915-019-0695-8
—————————————-
Scarless whole-body regeneration in the absence of a blastema requires cell division in the ctenophore Mnemiopsis leidyi
2019
https://www.biorxiv.org/content/10.1101/509331v1.full.pdf
—————————————-
Transdifferentiation is a driving force of regeneration in Halisarca dujardini (Demospongiae, Porifera)
August 25, 2015
https://peerj.com/articles/1211/
—————————————-
Oscarella lobularis (Homoscleromorpha, Porifera) Regeneration: Epithelial Morphogenesis and Metaplasia
August 13, 2015
https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0134566
—————————————-
Reconsidering regeneration in metazoans: an evo-devo approach
https://www.frontiersin.org/articles/10.3389/fevo.2015.00067/full
—————————————-
Regeneration in the metazoans: why does it happen?
2000
https://pubmed.ncbi.nlm.nih.gov/10842312/
—————————————-
Regeneration across metazoan phylogeny: lessons from model organisms.
20 February 2015
https://www.semanticscholar.org/paper/Regeneration-across-metazoan-phylogeny%3A-lessons-Li-Yang/95ca6cefc8032bafd53f5572e0213d02f04d0b36
—————————————-
Metazoan
https://www.sciencedirect.com/topics/earth-and-planetary-sciences/metazoan
—————————————-
Hydra as a unique model for the study of regenerative mechanisms in metazoans
September 16, 2019
https://medcraveonline.com/MOJAP/hydra-as-a-unique-model-for-the-study-of-regenerative-mechanisms-in-metazoans.html
—————————————-
The cAMP response element binding protein (CREB) as an integrative HUB selector in metazoans: clues from the hydra model system
2007
https://archive-ouverte.unige.ch/unige:13151
—————————————-
JmjC Domain-Encoding Genes Are Conserved in Highly Regenerative Metazoans and Are Associated with Planarian Whole-Body Regeneration.
2019
https://europepmc.org/article/PMC/PMC6390904
—————————————-
Identification of genes needed for regeneration, stem cell function, and tissue homeostasis by systematic gene perturbation in planaria
2005
https://www.ncbi.nlm.nih.gov/labs/pmc/articles/PMC2267917/
—————————————-
The Akt signaling pathway is required for tissue maintenance and regeneration in planarians
11 April 2016
https://bmcdevbiol.biomedcentral.com/articles/10.1186/s12861-016-0107-z
—————————————-
Small RNAs regulate stem cell function and regeneration in the planarian
January 2010
https://opencommons.uconn.edu/dissertations/AAI3420192/
—————————————-
ERK signaling controls blastema cell differentiation during planarian regeneration
2011
https://journals.biologists.com/dev/article/138/12/2417/44442/ERK-signaling-controls-blastema-cell
—————————————-
A molecular wound response program associated with regeneration initiation in planarians
2012
http://genesdev.cshlp.org/content/26/9/988
—————————————-
Germ cell specification and regeneration in planarians
2008
https://planosphere.stowers.org/pub/137
—————————————-
Germline Regeneration: The Worms’ Turn
2006
https://www.academia.edu/64888367/Germline_Regeneration_The_Worms_Turn
—————————————-
Key gene in early brain regeneration in planarians described by scientists
May 19, 2014
https://www.sciencedaily.com/releases/2014/05/140519105810.htm
—————————————-
Planarian Regeneration: Its End Is Its Beginning
2006
https://www.cell.com/fulltext/S0092-8674(06)00060-2
—————————————-
Dying Young as Late as Possible: Planarians, Regeneration and Stem Cells
September 3, 2008
https://videocast.nih.gov/watch=6984
—————————————-
Planaria
Planaria is a genus of planarians in the family Planariidae. When an individual is cut into pieces, each piece has the ability to regenerate into a fully formed individual.
https://en.wikipedia.org/wiki/Planaria
—————————————-
Retardation of Planaria Regeneration by Ultraviolet Radiation
2003
https://www.jstor.org/stable/4608683
—————————————-
Is the Germline Immortal and Continuous? A Discussion in Light of iPSCs and Germline Regeneration
September 4, 2019
https://zenodo.org/record/3385322#.YfCoLPhME2x
—————————————-
Evolutionary origins of germline segregation in Metazoa: evidence for a germ stem cell lineage in the coral Orbicella faveolata (Cnidaria, Anthozoa)
13 January 2016
https://royalsocietypublishing.org/doi/10.1098/rspb.2015.2128
—————————————-
The roles and activation of endocardial Notch signaling in heart regeneration
01 February 2021
Abstract
As a highly conserved signaling pathway in metazoans, the Notch pathway plays important roles in embryonic development and tissue regeneration. Recently, cardiac injury and regeneration have become an increasingly popular topic for biomedical research, and Notch signaling has been shown to exert crucial functions during heart regeneration as well. In this review, we briefly summarize the molecular functions of the endocardial Notch pathway in several cardiac injury and stress models. Although there is an increase in appreciating the importance of endocardial Notch signaling in heart regeneration, the mechanism of its activation is not fully understood. This review highlights recent findings on the activation of the endocardial Notch pathway by hemodynamic blood flow change in larval zebrafish ventricle after partial ablation, a process involving primary cilia, mechanosensitive ion channel Trpv4 and mechanosensitive transcription factor Klf2.
https://cellregeneration.springeropen.com/articles/10.1186/s13619-020-00060-6
—————————————-
Notch Signalling: The Multitask Manager of Inner Ear Development and Regeneration
15 February 2020
https://link.springer.com/chapter/10.1007/978-3-030-34436-8_8
—————————————-
Regeneration and the need for simpler model organisms
16 April 2004
https://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.519.6802&rep=rep1&type=pdf
—————————————-
The cellular basis for animal regeneration
2011
https://www.ncbi.nlm.nih.gov/labs/pmc/articles/PMC3139400/
—————————————-
Finding Common Ground in the Regeneration Process
February 20, 2019
https://www.cmu.edu/bio/news/2019/hinman_bmcbiology.html
—————————————-
The Diversity of Muscles and Their Regenerative Potential across Animals
2020
https://moh-it.pure.elsevier.com/en/publications/the-diversity-of-muscles-and-their-regenerative-potential-across-
—————————————-
Animal regeneration in the era of transcriptomics
6 Oct 2021
https://hal.archives-ouvertes.fr/hal-03153122/document
—————————————-
Retina repair, stem cells and beyond
2004
https://pubmed.ncbi.nlm.nih.gov/16181073/
—————————————-
Axonal regeneration of retinal ganglion cells after optic nerve pre-lesions and attachment of normal or pre-degenerated peripheral nerve grafts
12 November 2002
https://pubmed.ncbi.nlm.nih.gov/12507332/
—————————————-
Beyond stem cells, regenerative medicine finds exosomes
January 24, 2022
https://mcpress.mayoclinic.org/research-innovation/beyond-stem-cells-regenerative-medicine-finds-exosomes/
—————————————-
Nuclei-free cells prove utility in delivering therapeutics to diseased tissues
January 14, 2022
https://medicalxpress.com/news/2022-01-nuclei-free-cells-therapeutics-diseased-tissues.html
—————————————-
Wound signaling of regenerative cell reprogramming
2016
https://www.sciencedirect.com/science/article/abs/pii/S0168945216301315
—————————————-
Cell plasticity in homeostasis and regeneration.
02 July 2010
https://europepmc.org/article/MED/20602493
—————————————-
Piezoelectric Nano-Biomaterials for Biomedicine and Tissue Regeneration
18 February 2020
https://onlinelibrary.wiley.com/doi/10.1002/adfm.201909045
—————————————-
New wearable device turns the body into a battery
Feb. 10, 2021
https://www.colorado.edu/today/2021/02/10/thermoelectric
—————————————-
Electrospinning for healthcare: recent advancements
2020

Fig. 6 The use of polydopamine chemistry to facilitate cell growth on ES fibres. Scale bar: 100 μm. Fibres were prepared from pol(L-lactide) (PLLA), and then immersed in polydopamine to yield PLLA–PDA fibres. These were further immersed in a solution of the osteogenic growth peptide (OGP) to conjugate the peptide and generate PLLA–PDA–OGP. Cell nuclei are blue and the intracellular protein F-actin is green. The presence of OGP on the fibre surface clearly encourages cell growth. Reproduced from ref. 59 (Liu et al., 2019) under the terms of the Creative Commons CC BY 3.0 license. Published by The Royal Society of Chemistry.
—————————————-
How does the body make electricity — and how does it use it?
Apr 14, 2021
https://health.howstuffworks.com/human-body/systems/nervous-system/human-body-make-electricity.htm
—————————————-
How to Slow Aging (and even reverse it)
Dec 14, 2019
https://www.youtube.com/watch?v=QRt7LjqJ45k
—————————————-
Dr. Michio Kaku: “You will have the option to reach the age of 30 and stop”
Aug 17, 2021
https://www.youtube.com/watch?v=AL4QxWEpAI0
—————————————-
The Promise of Human Regeneration: Forever Young
Mar 23, 2018
https://www.youtube.com/watch?v=e0vKOYQUmgg
—————————————-
AKG human aging trials | Prof Brian Kennedy
Sep 7, 2021
https://www.youtube.com/watch?v=Z700Za1F0gY
—————————————-
Application of Stem Cell Technology in Antiaging and Aging-Related Diseases
2018
https://pubmed.ncbi.nlm.nih.gov/30232764/
—————————————-
Gene expressions related to DNA changes due to aging found to be related to CpG islands
December 22, 2021
https://medicalxpress.com/news/2021-12-gene-dna-due-aging-cpg.html
—————————————-
Study shows protein that reverses aging of skeletal muscle
September 14, 2021
https://medicalxpress.com/news/2021-09-protein-reverses-aging-skeletal-muscle.html
—————————————-
Downregulated miR-204 Promotes Skeletal Muscle Regeneration
17 Nov 2020
https://www.hindawi.com/journals/bmri/2020/3183296/
—————————————-
A new injectable hydrogel for cartilage repair
September 13, 2021
A team of researchers affiliated with a host of institutions in China has developed an injectable hydrogel for use in repairing damaged cartilage. In their paper published in the journal Science Advances, the group describes how they made their hydrogel, how it can be applied and how well it worked when tested on mice and pigs.
Repair of torn or eroded cartilage has improved dramatically in recent years as scientists have learned to grow chondrocytes (cells that grow into cartilage) and to use them to encourage growth of new cartilage. They are typically grown on structures in patches which are then applied to the area in need of repair. One major drawback to such treatment, however, is the need to cut through the skin and lay open the area to be treated. Such treatment can lead to a painful recovery over several months. In this new effort, the researchers have developed a type of hydrogel that can be used for the same type of treatment without the need for surgery.
The hydrogel developed by the team involved using light-initiated polymerization as well as light-induced cross-linked imine organic compounds. The result was a gel that could be applied to a scaffold and which would harden only when exposed to ultraviolet light. This meant that a scaffold could be rolled into a very small shape and inserted by injection to the site needing repair. The gel could then be injected onto the scaffolding. Once in place, all that was needed for the gel to harden in place was to shine a UV light onto the impacted area. The gel would harden within ten seconds allowing the scaffolding with its load of chondrocytes to grow new cartilage.
The researchers tried a variety of scaffolding shapes using lab mice and finally settled on a star-shape. They then further tested their gel by injecting several mice and allowing cartilage to grow for eight weeks. They next tested their approach using pigs that had defective cartilage and monitored the growth of cartilage using MRI scans. They found that after approximately six months, the cartilage in the pigs was restored.
The researchers plan to continue testing their hydrogel approach and expect to begin human trials soon—if all turns out well, they expect their approach to become standard treatment for cartilage repair.
https://medicalxpress.com/news/2021-09-hydrogel-cartilage.html
—————————————-
3-D Bioprinting in cartilage tissue engineering for bioinks-short review
2021
https://www.sciencedirect.com/science/article/pii/S2214785321042528
—————————————-
A novel therapeutic strategy for cartilage diseases based on lipid nanoparticle-RNAi delivery system
2018
https://pubmed.ncbi.nlm.nih.gov/29440889/
—————————————-
Bioactive Scaffolds for Regeneration of Cartilage and Subchondral Bone Interface
2018
https://www.thno.org/v08p1940.htm
—————————————-
Fountain of youth for the epigenome could become important in the treatment of osteoporosis
Sep 15 2021
https://www.news-medical.net/news/20210915/Fountain-of-youth-for-the-epigenome-could-become-important-in-the-treatment-of-osteoporosis.aspx
—————————————–
The Different Types of Stem Cell Therapy in Treating Osteoarthritis
2020
The main types of stem cell therapy are:
Bone Marrow derived stem cells. There are cells taken from your own bone marrow and injected into problem areas.
Adipose derived stem cells. There are cells taken from your own adipose or fat cells and injected into problem areas. The fat is taken typically from your abdominal area in a liposuction type procedure.
Umbilical cord stem cells and placenta derived stem cells. These are cells taken from the afterbirth umbilical cord blood and placenta material donated by the new birth mothers. These mothers donate the afterbirth materials to laboratories, after a screening process, the laboratories make these cells available available to us for use in our patients. The donation does not harm either baby or mother in anyway. These cells are tested for infectious disease and the laboratories send us those testing results when we order the product.
—————————————–
Researchers may have discovered fountain of youth by reversing aging in human cells
May 27, 2015
http://www.gizmag.com/reversal-of-aging-human-cell-lines/37721/
Researchers in Japan have found that human aging may be able to be delayed or even reversed, at least at the most basic level of human cell lines. In the process, the scientists from the University of Tsukuba also found that regulation of two genes is related to how we age.
The new findings challenge one of the current popular theories of aging, that lays the blame for humans’ inevitable downhill slide with mutations that accumulate in our mitochondrial DNA over time. Mitochondrion are sometimes likened to a cellular “furnace” that produces energy through cellular respiration. Damage to the mitochondrial DNA results in changes or mutations in the DNA sequence that build up and are associated with familiar signs of aging like hair loss, osteoporosis and, of course, reduced lifespan.
So goes the theory, at least. But the Tsukuba researchers suggest that something else may be going on within our cells. Their research indicates that the issue may not be that mitochondrial DNA become damaged, but rather that genes get turned “off” or “on” over time. Most intriguing, the team led by Professor Jun-Ichi Hayashi was able to flip the switches on a few genes back to their youthful position, effectively reversing the aging process.
The researchers came to this conclusion by comparing the function level of the mitochondria in fibroblast cell lines from children under 12 years of age to those of elderly people between 80 and 97. As expected, the older cells had reduced cellular respiration, but the older cells did not show more DNA damage than those from children. This discovery led the team to propose that the reduced cellular function is tied to epigenetic regulation, changes that alter the physical structure of DNA without affecting the DNA sequence itself, causing genes to be turned on or off. Unlike mutations that damage that sequence, as in the other, aforementioned theory of aging, epigenetic changes could possibly be reversed by genetically reprogramming cells to an embryonic stem cell-like state, effectively turning back the clock on aging.
For a broad comparison, imagine that a power surge hits your home’s electrical system. If not properly wired, irreversible damage or even fire may result. However, imagine another home in which the same surge trips a switch in this home’s circuit breaker box. Simply flipping that breaker back to the “on” position should make it operate as good as new. In essence, the Tsukuba team is proposing that our DNA may not become fried with age as previously thought, but rather simply requires someone to access its genetic breaker box to reverse aging.
To test the theory, the researchers found two genes associated with mitochondrial function and essentially experimented with turning them on or off. In doing so, they were able to create defects or restore cellular respiration. These two genes regulate glycine, an amino acid, production in mitochondria, and in one of the more promising findings, a 97-year-old cell line saw its cellular respiration restored after the addition of glycine for 10 days.
The researchers’ findings were published this month in the journal Scientific Reports.
Whether or not this process could be a potential fountain of youth for humans and not just human fibroblast cell lines still remains to be seen, with much more testing required. However, if the theory holds, glycine supplements could one day become a powerful tool for life extension.
—————————————–
A barrier against brain stem cell aging
September 17, 2015
Neural stem cells generate new neurons throughout life in the mammalian brain. However, with advancing age the potential for regeneration in the brain dramatically declines. Scientists of the University of Zurich now identified a novel mechanism of how neural stem cells stay relatively free of aging-induced damage. A diffusion barrier regulates the sorting of damaged proteins during cell division.
http://phys.org/news/2015-09-barrier-brain-stem-cell-aging.html#jCp
—————————————–
Microbes turn back the clock as research discovers their potential to reverse aging in the brain
August 9, 2021
https://www.sciencedaily.com/releases/2021/08/210809122202.htm
—————————————–
Poop Transplants From Youngsters Reverse Brain Aging In Mice
2021
https://www.iflscience.com/health-and-medicine/poop-transplants-from-youngsters-reverse-brain-aging-in-mice/
—————————————–
Mouse and human germline cells appear to reset their biological age
June 30, 2021
https://medicalxpress.com/news/2021-06-mouse-human-germline-cells-reset.html
—————————————–
Researchers discover surprisingly wide variation across species in genetic systems that influence aging
May 28th, 2015
http://phys.org/news/2015-05-surprisingly-wide-variation-species-genetic.html
A new Iowa State University study focusing on insulin signaling uncovered surprising genetic diversity across reptiles, birds and mammals.
The research sets the stage for an improved understanding of metabolism, growth and aging and may have implications for medicine and human health, said Anne Bronikowski, an associate professor of ecology, evolution and organismal biology and a lead author of the study.
Insulin signaling is a critical biological process that governs the rate at which cells grow and divide and ultimately regulates aging. Scientists previously assumed the process remained much the same throughout the animal kingdom. But the new research shows that the genetic pathways in reptiles evolved to include protein forms not observed in mammals, a finding that suggestions these proteins carry out new or additional functions in reptiles.
The researchers looked at a molecular network known as the insulin/insulin-like signaling and target of rapamycin network (IIS/TOR). Because the IIS/TOR network regulates critical aspects of animal biology, scientists have long speculated that the network would work more or less the same in most animal species.
Bronikowski and Fred Janzen, a professor of ecology, evolution and organismal biology, completed the study along with former and current members of their labs. The research team compared the genomes of mammals and birds with 17 reptile species. The team found an abundance of variation in the hormones and receptors of the network, which bucks the conventional wisdom and indicates that hormones delivered through insulin likely undertake additional functions in reptiles.
“The study provides a critical step toward understanding how the IIS/TOR network may regulate variation in metabolism, modes of reproduction and rates of aging,” Bronikowski said. “It highlights genetic variants that occur in nature that may be useful in a human health context. It therefore lays the groundwork for future research to identify natural genetic variants that may work together to alter the function of this network, which may lend insight into metabolic and aging diseases and treatments.”
Previous studies of insulin signaling have focused on species commonly used as laboratory models, such as mice, fruit flies and nematodes. The new study compared 66 species, including 17 reptile species for which the research group had to generate transcriptome data – or the set of all RNA molecules in a particular genome – because the data didn’t exist previously. The team was able to sequence the reptile data with the help of support from the ISU Center for Integrated Animal Genomics.
The wide range of variation discovered by the study may suggest that the insulin signaling network could be targeted by new medical therapies to treat conditions associated with aging, Bronikowski said.
—————————————————-
“Avatar” project aims for human immortality by 2045
July 25, 2012
http://www.gizmag.com/avatar-project-2045/23454/
Russian media magnate Dmitry Itskov is heading “Avatar,” a tremendously ambitious and far-reaching multidisciplinary research project that aims to achieve immortality in humans within the next three decades. He plans to do it by housing human brains in progressively more disembodied vehicles, first transplanting them into robots and then, by the year 2045, by reverse-engineering the human brain and effectively “downloading” human consciousness onto a computer chip.
Fact or fiction?
When speculating on seemingly unobtainable goals such as this, one must be careful not to believe that improbable technological advances automatically become more likely simply by looking further away in the future. This is the cognitive trap that, for instance, has seen many leading IT experts predict the development of a human-level artificial intelligence at roughly twenty years in the future for at least the past five decades
———————————————
Can We Reprogram Our DNA and Heal Ourselves With Frequency, Vibration & Energy?
March 5, 2014
http://www.wakingtimes.com/2014/03/05/reprogram-dna-heal-ourselves-frequency-vibration-energy/
————————————————–
Novel UV-mediated mode of DNA repair
December 22, 2015
UV light damages DNA. But LMU researchers now show that it can also mediate non-enzymatic repair of one type of damage, albeit in a specific context. This effect may have played vital role in early evolution of living systems.The ultraviolet fraction of sunlight triggers photochemical reactions in the DNA molecules that make up the hereditary material in our cells. These reactions can result in alterations in the DNA structure which lead to mutations that may cause cell death or promote tumorigenesis. LMU researchers led by Professor Thomas Carell, who holds the Chair of Bioorganic Chemistry, and Wolfgang Zinth, Professor of Biomolecular Optics, have now shown that the DNA itself can repair one of the most common types of UV-mediated damage by a non-enzymatic mechanism which is itself dependent on UV. The new findings appear in the Journal of the American Chemical Society.
Cells possess a variety of complex, enzyme-based mechanisms which are used for the repair of damaged DNA, and this year’s Nobel Prize in Chemistry was awarded to three researchers for work on the elucidation of these mechanisms. The team led by Zinth and Carell, have now discovered the first instance of a sequence-dependent repair mechanism which does not require the participation of enzymes at all.
http://phys.org/news/2015-12-uv-mediated-mode-dna.html
———————————————-
Scientists Prove DNA Can Be Reprogrammed by Words and Frequencies
Aug 2, 2012
THE HUMAN DNA IS A BIOLOGICAL INTERNET and superior in many aspects to the artificial one. Russian scientific research directly or indirectly explains phenomena such as clairvoyance, intuition, spontaneous and remote acts of healing, self healing, affirmation techniques, unusual light/auras around people (namely spiritual masters), mind’s influence on weather patterns and much more. In addition, there is evidence for a whole new type of medicine in which DNA can be influenced and reprogrammed by words and frequencies WITHOUT cutting out and replacing single genes.
http://wakeup-world.com/2011/07/12/scientist-prove-dna-can-be-reprogrammed-by-words-frequencies/
———————————————–
The future of medicine could be found in this tiny crystal ball
February 4, 2016
A Drexel University materials scientist has discovered a way to grow a crystal ball in a lab. Not the kind that soothsayers use to predict the future, but a microscopic version that could be used to encapsulate medication in a way that would allow it to deliver its curative payload more effectively inside the body.

———————————————–
Doctors Try Genetically Modified Poliovirus As Experimental Brain Cancer Treatment
2018
https://www.npr.org/sections/health-shots/2018/06/26/622610333/doctors-try-genetically-modified-poliovirus-as-experimental-brain-cancer-treatme
A genetically modified poliovirus may help some patients fight a deadly form of brain cancer, researchers report.
The experimental treatment seems to have extended survival in a small group of patients with glioblastoma who faced a grim prognosis because standard treatments had failed, Duke University researchers say.
“I’ve been doing this for 50 years and I’ve never seen results like this,” says Dr. Darell Bigner, the director emeritus of the The Preston Robert Tisch Brain Tumor Center at the Duke Cancer Institute, who is helping develop the treatment.

(Poliovirus, long a scourge, has been modified by Duke University researchers for experimental use as a brain cancer treatment).
————————————
Clinical Trial Studies Treating Brain Tumor with Genetically Modified Poliovirus
January 27, 2020
https://www.uhhospitals.org/Healthy-at-UH/articles/2020/01/clinical-trial-studies-treating-brain-tumor-with-genetically-modified-poliovirus
————————————
Poliovirus and Rhinovirus
https://pdb101.rcsb.org/motm/020
————————————
Vesicular stomatitis virus as a flexible platform for oncolytic virotherapy against cancer
2012
https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4091291/
—————————–
COVID-19 May Manifest in the Mouth in Some Patients — Pay attention to those with a rash, small case series from Spain advises
7-15-2020
https://www.medpagetoday.com/infectiousdisease/covid19/87584
You can add enanthem, or a rash inside the body, such as in the mouth, to the ever-lengthening list of symptoms with which COVID-19 patients can present, researchers in Spain suggested.
Six of 21 patients with an exterior skin rash also had these lesions, which seemed to appear about 2 weeks after symptom onset, reported Juan Jimenez-Cauhe, MD, of Hospital Universitario Ramon y Cajal in Madrid, and colleagues, in a research letter in JAMA Dermatology.
—————————–
COVID toes, rashes: How the coronavirus can affect your skin
https://www.aad.org/public/diseases/coronavirus/covid-toes
—————————–
9 Things Your Feet Can Tell You About Your Liver Problems – Dr.Berg
Dec 29, 2018
https://www.youtube.com/watch?v=CsTmgX32Uk8
—————————–
8 Types of Rashes That Can Be a Sign of COVID-19
March 18, 2021
https://www.health.com/condition/infectious-diseases/coronavirus/types-of-rashes-covid-19
One study found that a rash was the only symptom of COVID for 21% of patients.
Neck & exposed chest eczema
Oral rash
Papular & vesicular rashes
Pityriasis rosea
Purpuric or vasculitic rashes
Urticarial rash
Viral exanthem
—————————–
Red Spots on Legs Causes, Pictures Including Small, Itchy, Non Itchy Dots on Legs
July 5, 2021
https://americanceliac.org/red-spots-on-legs-causes-pictures-dots/
—————————–
Dark Spots on Legs, Causes, Lower, Thighs, Bruises, Shaving, Cream, Removal, Get Rid
August 16, 2017
https://healthrid.org/dark-spots-on-legs-causes-cancer-cream-removal/
—————————–
Sunlight Can Wake Up the Latent Herpes Virus; polyDNA Surveys the Public Knowledge
May 23, 2013
https://www.prweb.com/releases/2013/5/prweb10758945.htm
—————————–
Can You Go in the Sun with Shingles?
June 18, 2020
https://blogs.webmd.com/public-health/20200618/can-you-go-in-the-sun-with-shingles
—————————–
Study: Some COVID vaccines may reactivate shingles or herpes zoster
Apr 21, 2021
https://fox2now.com/news/national/study-some-covid-vaccines-may-reactivate-shingles-or-herpes-zoster/
ST. LOUIS, Mo. – A new study finds a link between COVID vaccine and a skin rash. The Rheumatology journal reports that some patients developed herpes zoster, also known as shingles, after receiving the Pfizer vaccine.
Only six people out of the 491 patients in the study experienced the infection, researchers said. They had mild cases.
The viral zoster infection may cause a painful rash. It is most commonly found as a stripe of blisters on the side of your torso. It is caused by the same virus that causes chickenpox. People with autoimmune inflammatory rheumatic diseases have a higher risk of developing the herpes infection.
The CDC estimates that one million cases of herpes zoster occur annually in the United States. Herpes zoster rates are increasing among adults in the United States, especially among younger adults. The increase has been gradual over a long period of time.
The researchers conducting this study say that there appears to be a link between mRNA vaccines, people with autoimmune inflammatory diseases, and the reactivation of the zoster virus. They say that more studies should be conducted to determine the link.
—————————–
A Herpes Infection May Be Linked to the COVID Vaccine—Here’s Why You Shouldn’t Panic
April 21, 2021
We asked experts to break down the results of a small new study, which found that herpes zoster might be a side effect of getting vaccinated.
https://www.health.com/condition/infectious-diseases/coronavirus/herpes-covid-vaccine
—————————–
Study: Shingles may be side effect of COVID-19 vaccine
April 20, 2021
https://whdh.com/news/study-shingles-may-be-side-effect-of-covid-19-vaccine/
—————————–
Experts Debunk COVID-19 Vaccine-Shingles Link
Feb. 9, 2021
https://www.webmd.com/vaccines/covid-19-vaccine/news/20210209/experts-debunk-covid-19-vaccine-shingles-link
—————————–
Reports of rashes and skin reactions after COVID vaccine
4/7/2021
https://www.msn.com/en-us/health/medical/reports-of-rashes-and-skin-reactions-after-covid-vaccine/ar-BB1fpn3N
—————————–
The COVID-19 Vaccine Can Cause Delayed Skin Reactions—Here’s Everything We Know
Mar 4, 2021
https://www.newbeauty.com/covid-vaccine-skin-reactions/
—————————–
Possible Side Effects After Getting a COVID-19 Vaccine
June 24, 2021
https://www.cdc.gov/coronavirus/2019-ncov/vaccines/expect/after.html
—————————–
Doctors found a weird new coronavirus vaccine side effect that takes a week to appear
3/4/2021
https://www.msn.com/en-us/health/medical/doctors-found-a-weird-new-coronavirus-vaccine-side-effect-that-takes-a-week-to-appear/ar-BB1efwny
—————————–
Here’s why some are experiencing side effects after 2nd Pfizer or Moderna vaccine shot
April 19, 2021
Also: “What’s the difference between a normal reaction to the vaccine and a reaction that you should be concerned about?”
https://abc7ny.com/covid-19-vaccine-side-effects-pfizer-moderna-covid-19/10521642/
—————————–
5 people share their Pfizer and Moderna vaccine side effects, from arm pain to aches and fevers
Mar 11, 2021
https://www.businessinsider.com/covid-19-vaccine-side-effects-moderna-pfizer-2021-2?op=1
—————————–
If This Happens After Your COVID Vaccine, it’s Not Normal, Say Experts
“Allergic reactions can be lifethreatening,” says the Mayo Clinic.
March 31, 2021
https://www.eatthis.com/news-covid-vaccine-side-effects-mayo-clinic/
—————————–
Doctors Are Warning You to “Be Prepared” for This After Your Second Dose
February 27, 2021
https://bestlifeonline.com/news-vaccine-side-effect/
This is why the second dose of the COVID vaccine can come with more side effects.
The most commonly reported side effects following COVID vaccination are pain or swelling at the injection site, chills, headache, fever, and fatigue, according to the CDC. The bright side? Most people who experience these symptoms say that their reactions have been mild to moderate, and are made better with the help of over-the-counter medications like ibuprofen or acetaminophen. And for the latest COVID news delivered straight to your inbox, sign up for our daily newsletter.
—————————–
‘Muscle pains, fatigue, headaches’: The side-effects of coronavirus vaccine
Dec 10, 2020
Both Pfizer and Moderna have admitted that taking their vaccines could result in side-effects similar to mild Covid symptoms such as muscle pain, chills and a headache
https://www.businesstoday.in/industry/pharma/story/muscle-pains-fatigue-headaches-the-side-effects-of-coronavirus-vaccine-281074-2020-12-10
—————————–
Dr. Fauci Says These 2 Side Effects Mean Your COVID Vaccine Is Working
February 2, 2021
While COVID vaccine side effects can seem alarming, medical experts have cautioned that they are typically mild to moderate, only last up to 48 hours, and are actually an indication that your shot is working. During a Jan. 28 interview with MSNBC, chief White House COVID adviser Anthony Fauci, MD, recently name dropped two side effects in particular that he feels should be viewed as a welcome sign that your immune system is responding to your vaccination…
4. Joint pain
3. Headache
2. Fatigue
1. Pain at the injection site
https://bestlifeonline.com/news-vaccine-side-effects/
—————————–
Dr. Fauci reveals which medications you shouldn’t take before your coronavirus vaccine
March 4th, 2021
https://bgr.com/science/covid-vaccine-cdc-safety-fauci-advil-tylenol/
As the COVID vaccination effort in the US continues to pick up steam, Dr. Anthony Fauci and the CDC have recently cautioned that anyone preparing to get vaccinated shouldn’t take pain relievers like Advil, Motrin, or Tylenol before their appointment. The underlying rationale is that these medications might prevent COVID vaccines from working exactly as intended. While there’s no conclusive proof that this is the case with the vaccines from Pfizer and Moderna, the CDC and Fauci are naturally erring on the side of caution.
“There are data in the vaccine literature, long predating COVID-19 and almost all [done] in children, that premedication with [fever-reducing drugs] like acetaminophen or ibuprofen decrease the antibody response to the first dose of vaccine,” Dr. David Cennimo told the Miami Herald a few weeks ago.
Commenting on the issue, Dr. Fauci recently said:
The mixed advice is based on the fact that there’s very little data on that. I mean, if you’re going to take something that suppresses an immunological response, then obviously, you don’t want to take something like that.
Something that’s a true anti-inflammatory, such as one of the nonsteroidal anti-inflammatories, should not be given.
Consequently, the CDC currently advises against taking medication before getting vaccinated if the sole objective is to try and prevent side effects from manifesting.
However, if you’re already taking a medication like Tylenol for an existing medical condition as directed by your doctor, health experts say you should maintain the regimen.
It’s also worth noting that taking Advil of Tylenol post-vaccination is perfectly acceptable, with the CDC noting:
Antipyretic or analgesic medications (e.g., acetaminophen, non-steroidal anti-inflammatory drugs) can be taken for the treatment of post-vaccination local or systemic symptoms, if medically appropriate. However, routine prophylactic administration of these medications for the purpose of preventing post-vaccination symptoms is not currently recommended, because information on the impact of such use on mRNA COVID-19 vaccine-induced antibody responses is not available at this time.
—————————–
Paracetamol
https://en.wikipedia.org/wiki/Paracetamol
Paracetamol, also known as acetaminophen,[a] is a medication used to treat fever and mild to moderate pain. At a standard dose, paracetamol only slightly decreases body temperature; it is inferior to ibuprofen in that respect, and the benefits of its use for fever are unclear. Paracetamol may relieve pain in acute mild migraine but only slightly in episodic tension headache. However, the aspirin/paracetamol/caffeine combination helps with both conditions where the pain is mild and is recommended as a first-line treatment for them. Paracetamol is effective for post-surgical pain, but it is inferior to ibuprofen. The paracetamol/ibuprofen combination provides further increase in potency and is superior to either drug alone. The pain relief paracetamol provides in osteoarthritis is small and clinically insignificant. The evidence in its favor for the use in low back pain, cancer pain and neuropathic pain is insufficient…
In the short term, common side effects of paracetamol are nausea and abdominal pain, and it seems to have tolerability similar to ibuprofen. Chronic consumption of paracetamol may result in a drop in hemoglobin level, indicating possible gastrointestinal bleeding, and abnormal liver function tests. There is a consistent association of increased mortality as well as cardiovascular (stroke, myocardial infarction), gastrointestinal (ulcers, bleeding) and renal adverse effects with taking higher dose of paracetamol. The drug may also increase the risk of developing hypertension. Elevated frequency of asthma and developmental and reproductive disorders is observed in the offspring of women with prolonged use of paracetamol during pregnancy, although whether paracetamol is the true cause of this increase is unclear. Some studies suggest that there is evidence for the association between paracetamol during pregnancy and autism spectrum disorder and attention deficit hyperactivity disorder, while making clear further research is required to establish any causal link, which has prompted some calls to limit its use in pregnancy to the lowest effective dosage for the shortest possible time.
The recommended maximum daily dose for an adult is three to four grams. Higher doses may lead to toxicity, including liver failure. Paracetamol poisoning is the foremost cause of acute liver failure in the Western world, and accounts for most drug overdoses in the United States, the United Kingdom, Australia, and New Zealand.
Paracetamol was first made in 1877 or possibly 1852. It is the most commonly used medication for pain and fever in both the United States and Europe. It is on the World Health Organization’s List of Essential Medicines. Paracetamol is available as a generic medication, with brand names including Tylenol and Panadol among others. In 2019, it was the 145th most commonly prescribed medication in the United States, with more than 4 million prescriptions.
—————————–
Use of acetaminophen (paracetamol) during pregnancy and the risk of autism spectrum disorder in the offspring
2016
https://pubmed.ncbi.nlm.nih.gov/26930528/
—————————–
Acetaminophen (paracetamol) and autism? Is there a connection?
Jun 12, 2017
https://www.youtube.com/watch?v=oD_opnk4nxU
—————————–
Association of Cord Plasma Biomarkers of In Utero Acetaminophen Exposure With Risk of Attention-Deficit/Hyperactivity Disorder and Autism Spectrum Disorder in Childhood
October 30, 2019
https://jamanetwork.com/journals/jamapsychiatry/fullarticle/2753512
—————————–
Acetaminophen exposure during early childhood probably induces autism: Not a conspiracy theory, but rather a critical review from scientists and clinicians at Duke and Harvard
2017
https://openforest.net/acetaminophen-exposure-during-early-childhood-probably-induces-autism/
—————————–
Use of acetaminophen (paracetamol) during pregnancy and the risk of autism spectrum disorder in the offspring
2016
https://pubmed.ncbi.nlm.nih.gov/26930528/
—————————–